New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
checkpoint: pipeline finishing early #823
Comments
|
Could you create a minimal (well, small enough to post here) cc @jmeppley |
|
I'm still having problems creating a reproducible minimal example, but I am still definitely running into this problem. I ran a pipeline that finished nearly at the end after a couple of days: but then when I try to restart snakemake to finish those last steps, snakemake just starts from the beginning 😞 |
|
It sounds like you are running up against two separate problems.
First the workflow isn't running to completion, and that's probably
connected to the checkpoint (whether or not it's the same bug or a
different one).
However, the workflow starting over from the beginning is likely a totally
different problem. In all my issues with checkpoints, I haven't had the
workflow start over from the beginning on a re-run. But I've often had
other workflows start over from the beginning because of something weird in
my Snakefile.
I suggest debugging the restart behavior separately. The next time it stops
without finishing, use --dry-run and --reason to figure out what is
triggering the full re-run.
…On Fri, Jan 15, 2021 at 1:02 AM Nick Youngblut ***@***.***> wrote:
I'm still having problems creating a reproducible minimal example, but I
am still definitely running into this problem. I ran a pipeline that
finished nearly at the end after a couple of days:
[Thu Jan 14 21:44:51 2021]
Finished job 67.
72 of 87 steps (83%) done
Removing temporary output file /ebio/abt3_scratch/nyoungblut/LLPRIMER_101310211274/blastx/hits.txt.
[Thu Jan 14 22:09:43 2021]
Finished job 64.
73 of 87 steps (84%) done
Complete log: /ebio/abt3_projects/software/dev/ll_pipelines/llprimer/.snakemake/log/2021-01-14T195420.533178.snakemake.log
Pipeline complete! Creating report...
but then when I try to restart snakemake to finish those last steps,
snakemake just starts from the beginning 😞
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#823 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AANDP3HVG66IDK5WM52DTWLS2AAATANCNFSM4VR7CGDA>
.
|
|
I think there may be an issue with |
|
Hello, I think I might have the same issue. I have a rule to demultiplex Illumina data and I don't know which otuput I expect (or the number of files at least). I use checkpoints to continue the analysis (fastqc, multiqc, etc) after the checkpoint. The problem is that it runs fine until the checkpoint,. Then it properly (as far as I can tell) recalculates the DAG and then it stops (3% complete but exit 0) so it didn't really run the downstream rules. If I restart the pipeline, it starts where it left and properly finishes the jobs. I guess I can just put it in production like this, but it is not ideal. I am using |
|
This bug still exists. I don't have it with |
|
Here's a minimal reproducible example. Tested in a clean conda environment with snakemake 6.8.0 about the commandThe command used to run this file removes the about the codeMost my 2 centsThis example somewhat compares to our (@Maarten-vd-Sande and I) use-case, in that it uses a similar input function downstream of the checkpoint (and only the rules using it are affected). However, our workflow's checkpoint has only one output. The issue is therefore probably wider, and my guess would be it is related to input functions using checkpoints. |
|
And here's a reproducible example matching our use-case! Runs with The input function from the previous example did not matter for this one. Instead it's piping (similar to #132)! EDIT: extended the example with |
|
I found one more edge case while making the above example (sorry!) the following code works fine: but this stops early: in this case, it seems the target rule cannot contain files coming directly from a checkpoint. EDIT: simplified. |
|
So this one: That sounds like it's just some overly aggressive avoiding of running rules. If you add a |
|
Ok, so the problem with the first MRE is as follows: The job for checkpoint a is executed and a1.txt and a2.txt are created. The job finishes, and the temp files that aren't needed are deleted. This is verified by checking the depends of all jobs for each file. Jobs for rules depending on checkpoints do not have output yet, so they aren't in the list, so files needed only by those rules get deleted. The reason why the first temp() works is that this file is actually added to the required files list as a stub. MRE 1 works fine if you pass |
The output of checkpoint rules may be needed by other rules, which, at this point have not had a chance to add their file requirements to their depending list.
- Can't have the b.txt in first text be temp and expect it present, it will correctly be deleted. - Add third MRE which is targeting more of a log issue
The main scheduler loop terminates if there are no more runnable jobs. This tests if there still are jobs that should be run, but that we cannot seem to get to. If so, print an error message, print the files affected, and exit 1. The latter is important so Snakemake can, as hotfix, be rerun in a loop until it's actually completed all tasks.
|
@epruesse and @siebrenf, could the second MRE in that list have anything to do with #1173, which was merged into v6.8.1? I ran the second test with both Snakemake v6.8.0 and v6.8.1. It failed on v6.8.0 but had the expected output on v6.8.1. |
|
I was able to reproduce the second MRE with v6.9.1. The failure is not deterministic works about 33% of the time. It could just be that you got (un)lucky in your test with v6.8.1? |
Yup, that was it! I ran it 15 more times. It failed on 7 of those. |
- Incorporates @epruesse's fix for MRE snakemake#1 - Adds a fix for MRE snakemake#2 - properly marks group jobs as finished - Some minor updates to tests
* add failing tests 823 * fix mistakes * black * Fix the first two MREs from #823. - Incorporates @epruesse's fix for MRE #1 - Adds a fix for MRE #2 - properly marks group jobs as finished - Some minor updates to tests * Fix tests on Windows * Skip MRE 2 from 823 on Windows due to `pipe()` output Co-authored-by: Maarten-vd-Sande <maartenvandersande@hotmail.com> Co-authored-by: Johannes Köster <johannes.koester@uni-due.de>
* add failing tests 823 * fix mistakes * black * Fix the first two MREs from snakemake#823. - Incorporates @epruesse's fix for MRE #1 - Adds a fix for MRE #2 - properly marks group jobs as finished - Some minor updates to tests * Fix tests on Windows * Skip MRE 2 from 823 on Windows due to `pipe()` output Co-authored-by: Maarten-vd-Sande <maartenvandersande@hotmail.com> Co-authored-by: Johannes Köster <johannes.koester@uni-due.de>
* add failing tests 823 * fix mistakes * black * Update tests from #823 MREs - Can't have the b.txt in first text be temp and expect it present, it will correctly be deleted. - Add third MRE which is targeting more of a log issue * Add error message printed if #823 is encountered, also exit 1 The main scheduler loop terminates if there are no more runnable jobs. This tests if there still are jobs that should be run, but that we cannot seem to get to. If so, print an error message, print the files affected, and exit 1. The latter is important so Snakemake can, as hotfix, be rerun in a loop until it's actually completed all tasks. * Fix checkpoint+temp leads to incomplete run issue The aggressive early deletion of temp files must ignore checkpoints. Other rules depend on a checkpoint only via the first output file of the checkpoint (flagged string) as a stub. All other outputs marked as temp would be deleted (and have been before this patch). Since those files are missing, the dependent rules will never be runnable and the workflow ends before all targets have been built. * Don't break untiljobs * fix typo Co-authored-by: Maarten-vd-Sande <maartenvandersande@hotmail.com> Co-authored-by: Johannes Köster <johannes.koester@uni-due.de> Co-authored-by: Johannes Köster <johannes.koester@tu-dortmund.de>
|
Should be fixed by merging PR #1203. Thanks to all of you for the detective work, the fixes and the examples! |
|
After upgrading to v7.0.0, I've come across this error when using snakemake on a cluster. I'm still testing it some more, but it appears that when a job failed (because of a known/valid reason), when all jobs are done on the cluster, snakemake then exits with this error specifying that the job that failed on the cluster is a "remaining job". |
|
Came across this bug (?) in v7.3.0. Not sure why Is this really a bug? The message is confusing. @johanneskoester |
|
I also hit this, but in 7.3.8. There were upstream jobs that failed, and I had |
|
Ah yes, I think I hut it in the same situation and it sounds right, red herring. Probably new issue should be opened to deal with it. |
|
it seems that this problem happens randomly, most of my jobs were successfully finished, but only 4 of them went unfinished. |
|
I also had this in v7.8 last week, can someone comment on how to solve it? Is --keep-going responsible, and if so, how? I need --keep-going, cant remove it usually. |
|
@chrarnold You can use --keep-going safely. It's the same as with |
|
This is indeed a bug. A PR with a testcase that reproduces this would be very welcome. We think that at least one way to trigger this is by combining checkpoints with group jobs. |
|
Same bug occurred with ecoli test run: All jobs finished after restarting with --keep-going |
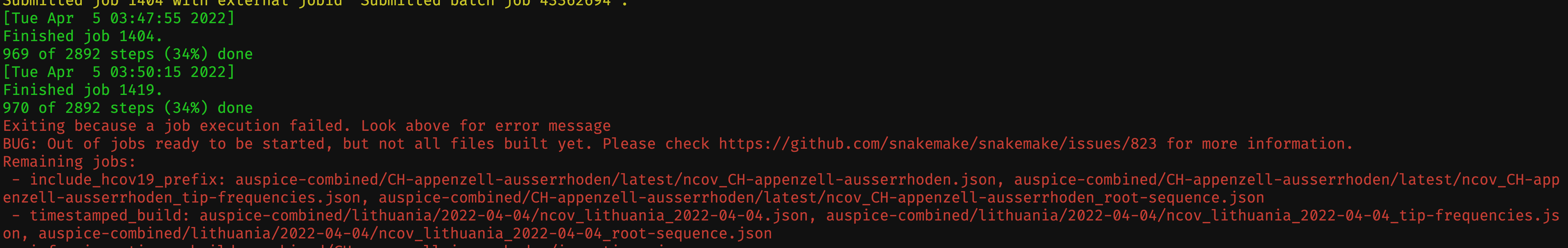
I'm running snakemake 5.31.1 (bioconda)
When using checkpoint, the pipeline will often finish successfully without completing all steps or even generating all output files designated by the
allrule.For instance, here's the end of a recent run:
If I try to "finish" the run by re-running snakemake, snakemake will start from the beginning of the pipeline instead of picking up where it left off.
I don't have a minimal example as of yet. Maybe others have (or will soon) run into this problem and post a minimal example from a simpler pipeline than mine.
The only checkpoint in my pipeline generates a large number of tsv files, and the output for that checkpoint rule is a directory. My aggregation function is:
I don't see why snakemake is ending the run early, especially since the final, aggregated output files designated in the
allrule were not created upon the early-finish.The text was updated successfully, but these errors were encountered: