New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[WIP] Feature names with pandas or xarray data structures #16772
[WIP] Feature names with pandas or xarray data structures #16772
Conversation
|
Thanks @thomasjpfan ! If you do benchmark, could you benchmark with pandas master as well please? |
|
Here is a benchmark for sparse data that runs the following script with different data = fetch_20newsgroups(subset='train')
def _run(*, max_features, array_out):
with config_context(array_out=array_out):
pipe = make_pipeline(CountVectorizer(max_features=max_features),
TfidfTransformer(),
SGDClassifier(random_state=42))
pipe.fit(data.data, data.target)Comparing between
|
|
Interesting. For xarray, you're returning a pydata/sparse array? Over in pandas-dev/pandas#33182, I (lightly) proposed accepting a pydata/sparse array in the DataFrame constructor. Since it implements the ndarray interface, we can store it in a 2D Block without much effort. Doing pretty much anything with it quickly breaks, since pandas calls Comparing scipy.sparse to pydata/sparse. sparse array -> DataFrame In [22]: shape = (100, 100_000)
In [23]: a = scipy.sparse.random(*shape)
In [24]: b = sparse.random(shape)
In [25]: a
Out[25]:
<100x100000 sparse matrix of type '<class 'numpy.float64'>'
with 100000 stored elements in COOrdinate format>
In [26]: b
Out[26]: <COO: shape=(100, 100000), dtype=float64, nnz=100000, fill_value=0.0>
In [27]: %timeit _ = pd.DataFrame.sparse.from_spmatrix(a)
1.12 s ± 24.3 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
In [28]: %timeit _ = pd.DataFrame(b)
6.84 ms ± 92.9 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)DataFrame -> sparse In [34]: df_scipy = pd.DataFrame.sparse.from_spmatrix(a)
In [35]: %timeit df_scipy.sparse.to_coo()
1.81 s ± 17.5 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
In [36]: df_sparse = pd.DataFrame(b)
In [37]: %timeit df_sparse.sparse.to_scipy_sparse()
549 µs ± 7.74 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)I would expect the memory usage to match xarray's (which IIUC is just the memory usage of the array + the index objects). |
The output of Although internally, the representation may get changed to another sparse format. For example, |
|
I didn't repeat the runs but it looks like there's no difference in peak memory for computing feature names using #12627: I added a call to pipe = make_pipeline(CountVectorizer(max_features=max_features),
TfidfTransformer(),
SGDClassifier(random_state=42))
pipe.fit(data.data, data.target)
if get_feature_names:
feature_names = pipe[:-1].get_feature_names() |
|
Btw, I would still like to see pandas in / pandas out. But I'm less certain about the sparse matrix support. So my current proposal would be to solve the 90% (95%?) usecase with the solution in #12627 and then add pandas in pandas out, and allow the |
|
@thomasjpfan have you checked if you see the same memory increase using pydata/sparse directly without xarray? That might help us narrow down the memory issue. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Btw, I would still like to see pandas in / pandas out. But I'm less certain about the sparse matrix support. So my current proposal would be to solve the 90% (95%?) usecase with the solution in #12627 and then add pandas in pandas out, and allow the
get_feature_namesin all cases.
I'd be 100% with you if we didn't have a pretty good solution already (this PR). Here's what I think:
- This PR is opt-in, and therefore it doesn't introduce any challenge to people who want to do mostly numerical work and are happy w/o the feature names which would be a main concern of @GaelVaroquaux
- This PR introduces a memory overhead proportionate to the number of columns (please correct me if I'm wrong @thomasjpfan) and therefore it wouldn't be an issue for common datasets.
- There's a significant overhead with very wide sparse data, e.g. NLP or any categorical data with many categories, which would mean users would not be able to use the feature names if they don't have the required memory, which means this PR doesn't satisfy everybody for now, but is still better than RFC Implement Pipeline get feature names #12627.
I would also argue that the overhead we see in xarray could probably be optimized if we figure it's an issue and there's enough demand from the community for the usecase to be better supported.
As somebody whose usecases are a part of that 5% @amueller mentions, I would really be much happier if we go down this route as I really see no blockers.
| @@ -59,6 +60,9 @@ def set_config(assume_finite=None, working_memory=None, | |||
|
|
|||
| .. versionadded:: 0.21 | |||
|
|
|||
| array_out : {'default', 'pandas', 'xarray'}, optional | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
should this be ndarray instead of default?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Sometimes the output is sparse. The default means "sparse or ndarray"
|
This PR has been updated to support In summary this PR is using following pattern to get the feature names into class MyEstimator(BaseEstimator):
def fit(self, X, y=None):
self._validate(X) # stores feature_names_in_ if avaliable
def transform(self, X, y=None):
X_orig = X
X = check_array(X)
# do the transform
return self._make_array_out(X, X_orig, get_feature_names_out='one_to_one')where For now This PR also implements SLEP 007 for feature names with column transformers and feature union. From an API point of view, if a library that uses scikit-learn and a user sets |
|
I think the third-party (or first-party) can protect against this by using a context manager (if we can make it threadsafe). This means that all third-party libraries need to wrap their code with a context manager if they want to guarantee a particular output. @adrinjalali suggests having a transform parameter (to overwrite the global). |
|
Not sure if this was clear before, but if we add the keyword/method and keep the global config, it's not very friendly to downstream libraries. They will have to branch on the version of sklearn. If we don't do a global config/context manager and only add the keyword/method then downstream libraries don't need to worry. |
|
If someone calls I'm not opposed to having an argument for |
|
would that enable the pipeline to pass around feature names down the path? |
|
This PR is on top of my personal wish list. @thomasjpfan and others: Many thanks! Status Quo
The last point with the global config option has 3 obstacles:
Alternatives mechanisms for controlling the ouput type are:
Next Steps = ConsolidationAre there news/solutions to obstacle point 2 parallelism? |
@adrinjalali To hopefully take a little burden away from this PR ... I think that many use cases requiring sparse data would be efficiently solved by providing native support for categorical data/columns, e.g. #16909. The data is often not sparse at all, only OHE makes it sparse and wide. |
|
Thank you for a great summary @lorentzenchr and I would also love to move forward. The "solution" for 2 is #17634, I think. long-term joblib might specify a mechanism to do that, but it would be up to the user to correctly use the mechanism.
I'm not entirely sure what you mean by that. I think if we want to move forward here, it might be ok for now to do the potentially silly but easy-to-understand thing of passing the output type to every step in the pipeline to simplify things. That might result in some extra wrapping and unwrapping but the user was very explicit that they want to work with a particular data type and if that's "slow" at first, that's not the worst that can happen. So moving forward could be:
I think #18010 might actually be a prerequisite for this, otherwise the output column names could be inconsistent, which seems a bit weird? The main motivation to go from the original A to B or C is that A puts more burden on downstream, no-one has thought about a thread-save context manager, and the joblib workaround in #17634 is a bit icky (but maybe also unavoidable for the working memory). Maybe a good way to decide this would be to first figure out if we need to have a thread-save context manager for A, and if so, how to do it. Then we can decide what the trade-off is between keyword and context managers? |
|
B and C have the issue that if we have a pipeline, and call A solution that I had discusses with @thomasjpfan that seems relatively reasonable is to do B/C and then also add a constructor argument to The benefit of having a keyword argument to |
|
Here's a proposal based on some of the above suggestions:
I'm not sure if I completely understand your solution @amueller |
|
@adrinjalali I don't understand how your solution helps downstream libraries. If a library uses sklearn and requires ndarray output, they have to inspect the signature of transform and then if Do you have a question about my solution lol? |
|
Right, I was thinking of the other way, as when third party library estimators are used in a sklearn meta estimator. So to understand your solution, would this be what you suggest?
If this is what you mean, I think it indeed covers most cases and I'm okay with it. |
|
Yes, that's about what I was thinking. It's a bit unclear what to do if Pipeline(..., transform_output_type='ndarray').transform(X, output_type='pandas')but maybe we just forbid that (or use ndarray in all but the last transform, which might give you useless feature names but whatever, the user asked for it). Generally I think |
|
@amueller could you please write an example where this would be an issue? |
|
Are the performance graphs at the top of this PR still valid? (ie up to date with the PR and the latest version of pandas or not much has changed)? If the performance loss is still that significant, I feel that we could suggest the following route:
|
I feel like this is will continue to be a blocker for After some thought, I wrote scikit-learn/enhancement_proposals#48 to propose something much simpler and will accomplish the almost the same goal as this PR. Furthermore, I see scikit-learn/enhancement_proposals#48 as almost a prerequisite for this PR:
|
|
Closing in favor of #20100 |
|
@thomasjpfan ,is this supported in scikit-learn now? I did check your link and seems that sklearn.set_config(array_out='') is still not supported. |
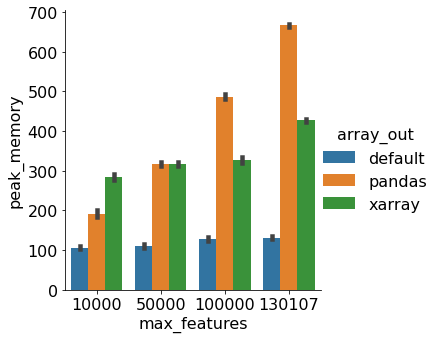



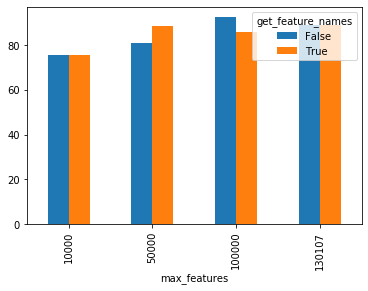
This is a prototype of what SLEP014 may look like with pandas/xarray + sparse support. The configuration flag,
array_out, only controls what comes out oftransform.This usage notebook contains how this API can be used with various transformers.
Updates
Notes
_DataAdapteris able to get the names of any thing that has a feature name. It can also wrap dense arrays and sparse matrix into apandas.DataFrameor axarray.DataArray.I am going to put together some benchmarks to to compare the following:
WIP: Feel free to look at the usage notebook. The internal implementation and its API is flux.