New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[WIP] Stable and fast float32 implementation of euclidean_distances #11271
[WIP] Stable and fast float32 implementation of euclidean_distances #11271
Conversation
82f16a1
to
ac5d1fb
Compare
|
Ping when you feel it's ready for review!
Thanks
|
|
@jnothman, Now that all tests pass, I think it's ready for review. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is certainly very precise, but perhaps not so elegant! I think it looks pretty good, but I'll have to look again later.
Could you report some benchmarks here please? Thanks
sklearn/metrics/pairwise.py
Outdated
| swap = False | ||
|
|
||
| # Maximum number of additional bytes to use to case X and Y to float64. | ||
| maxmem = 10*1024*1024 |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
please write down that this is 10MB
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Why do you believe this is a reasonable limit?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
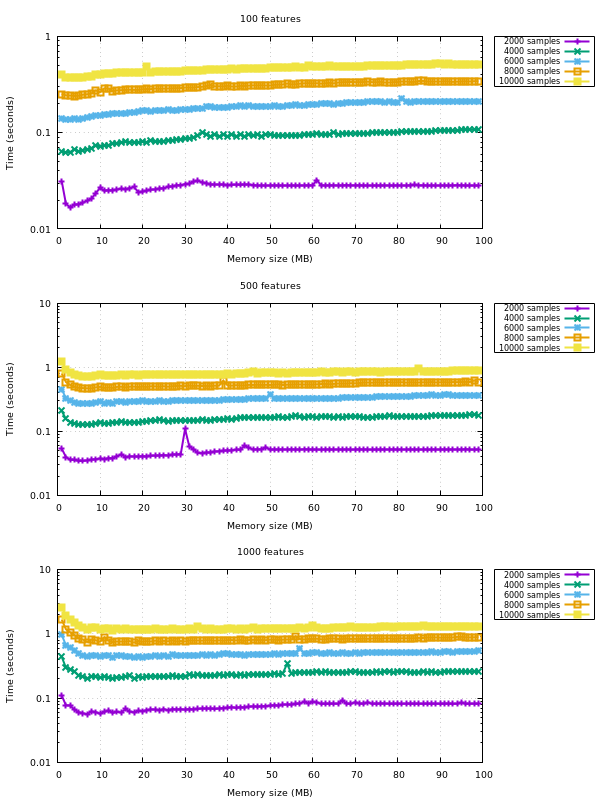
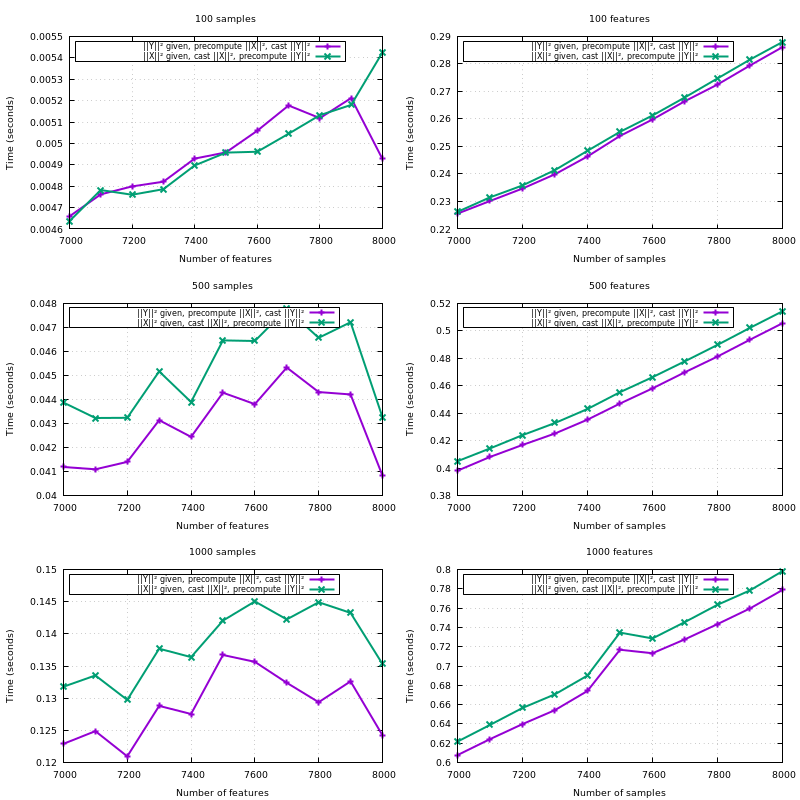
See the benchmark below that vary the memory size.
sklearn/metrics/pairwise.py
Outdated
| return distances if squared else np.sqrt(distances, out=distances) | ||
|
|
||
|
|
||
| def _cast_if_needed(arr, dtype): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
We'd usually put this sort of thing in sklearn.utils.fixes, which helps us find it for removal when our minimum dependencies change.
sklearn/metrics/pairwise.py
Outdated
| if X.dtype != np.float64: | ||
| XYmem += X.shape[1] | ||
| if Y.dtype != np.float64: | ||
| XYmem += Y.shape[1] |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Perhaps make this X.shape[1] just to reduce reader surprise since they should be equal...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
On the other hand, it's under a test for Y.dtype. So X.shape[1] would be equally surprising (at least to me). And this is to account for the memory occupied by the the float64 copy of Y. So semantically, it seems more appropriate to me to use Y.shape[1].
I might add a comment or something to make it more clear.
Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
c22b0bc
to
98e88d7
Compare
Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
…oat64 Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
Signed-off-by: Celelibi <celelibi@gmail.com>
98e88d7
to
0fd279a
Compare
I had to rebase my branch and force push it anyway for the auto-merge to succeed and the tests to pass. @jnothman wanna have a look at the benchmarks and discuss the mergability of those commits? |
|
yes, this got forgotten, I'll admit.
but I may not find time over the next week. ping if necessary
|
|
@rth, you may be interested in this PR, btw.
IMO, we broke euclidean distances for float32 in 0.19 (thinking that
avoiding the upcast was a good idea), and should prioritise fixing it.
|
|
Very nice benchmarks and PR @Celelibi ! It would be useful to also test the net effect of this PR on performance e.g. of KMeans / Birch as suggested in #10069 (comment) I'm not yet sure how this would interact with |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
A few more comments..
| # - a float64 copy of a block of X_norm_squared if needed; | ||
| # - a float64 copy of a block of Y_norm_squared if needed. | ||
| # This is a quadratic equation that we solve to compute the block size that | ||
| # would use maxmem bytes. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
What happens if we use a bit more? The performance is decreased?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The performance decreases a bit, yes. But it's only noticeable for a maxmem several order of magnitude larger than required. If you look at the first set of plots, you'll see a trend for larger memory sizes.
| distances[i:ipbs, j:jpbs] = d | ||
|
|
||
| if X is Y and j > i: | ||
| distances[j:jpbs, i:ipbs] = d.T |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Line not covered by tests, probably need to add a test for this
| if X_norm_squared is None and Y_norm_squared is not None: | ||
| X_norm_squared = Y_norm_squared.T | ||
| if X_norm_squared is not None and Y_norm_squared is None: | ||
| Y_norm_squared = X_norm_squared.T |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Also not covered by tests..
| D32_32 = euclidean_distances(X32, Y32) | ||
| assert_equal(D64_32.dtype, np.float64) | ||
| assert_equal(D32_64.dtype, np.float64) | ||
| assert_equal(D32_32.dtype, np.float32) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
assert D32_32.dtype == np.float32 etc.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
With the adoption of pytest, we are phasing out use of test helpers assert_equal, assert_true, etc. Please use bare assert statements, e.g. assert x == y, assert not x, etc.
| Y_norm_squared=Y32_norm_sq) | ||
| assert_greater(np.max(np.abs(DYN - D64)), .01) | ||
| assert_greater(np.max(np.abs(DXN - D64)), .01) | ||
| assert_greater(np.max(np.abs(DXYN - D64)), .01) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
assert np.max(np.abs(DXYN - D64)) > .01
| [0.9221151778782808, 0.9681831844652774]]) | ||
| D64 = euclidean_distances(X) | ||
| D32 = euclidean_distances(X.astype(np.float32)) | ||
| assert_array_almost_equal(D32, D64) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
On master I get,
In [5]: D64 = euclidean_distances(X)
In [6]: D64
Out[6]:
array([[0. , 0.00054379],
[0.00054379, 0. ]])
In [7]: D32 = euclidean_distances(X.astype(np.float32))
In [8]: D32
Out[8]:
array([[0. , 0.00059802],
[0.00059802, 0. ]], dtype=float32)and assert_array_almost_equal uses 6 decimals by default, so since this passes, it would increase the accuracy from 1 to 3 significant digits. Maybe use assert_allclose with rtol to make it more explicit?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This looks great for a start (i.e. as a bug fix, rather than an enhancement). I need to review some of your optimisations and benchmarks to understand whether it's worth enhancing it further.
| D32_32 = euclidean_distances(X32, Y32) | ||
| assert_equal(D64_32.dtype, np.float64) | ||
| assert_equal(D32_64.dtype, np.float64) | ||
| assert_equal(D32_32.dtype, np.float32) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
With the adoption of pytest, we are phasing out use of test helpers assert_equal, assert_true, etc. Please use bare assert statements, e.g. assert x == y, assert not x, etc.
I didn't forsee this. Well, they might chunk twice, which may have a bad impact on performance.
It wouldn't be easy to have the chunking done at only one place in the code. I mean the current code always use The best solution I could see right now would be to have some specialized chunked implementations for some metrics. Kind of the same way BTW
Interesting, I didn't know about that. However, it looks like
That comment was about deprecating |
|
I think given that we seem to see that this operation works well with 10MB
working mem and the default working memory is 1GB, we should consider this
a negligible addition, and not use the working_memory business with it.
I also think it's important to focus upon this as a bug fix, rather than
something that needs to be perfected in one shot.
|
|
@Celelibi Thanks for the detailed response!
@jeremiedbb, @ogrisel mentioned that you run some benchmarks demonstrating that using a smaller working memory had higher performance on your system. Would you be able to share those results (the original benchmarks are in #10280 (comment))? Maybe in a separate issue. Thanks! |
|
from my understanding this will take some more work. untagging 0.20. |
Reference Issues/PRs
Fixes #9354
Superseds PR #10069
What does this implement/fix? Explain your changes.
These commits implement a block-wise casting to float64 and uses the older code to compute the euclidean distance matrix on the blocks. This is done useing only a fixed amount of additional (temporary) memory.
Any other comments?
This code implements several optimizations:
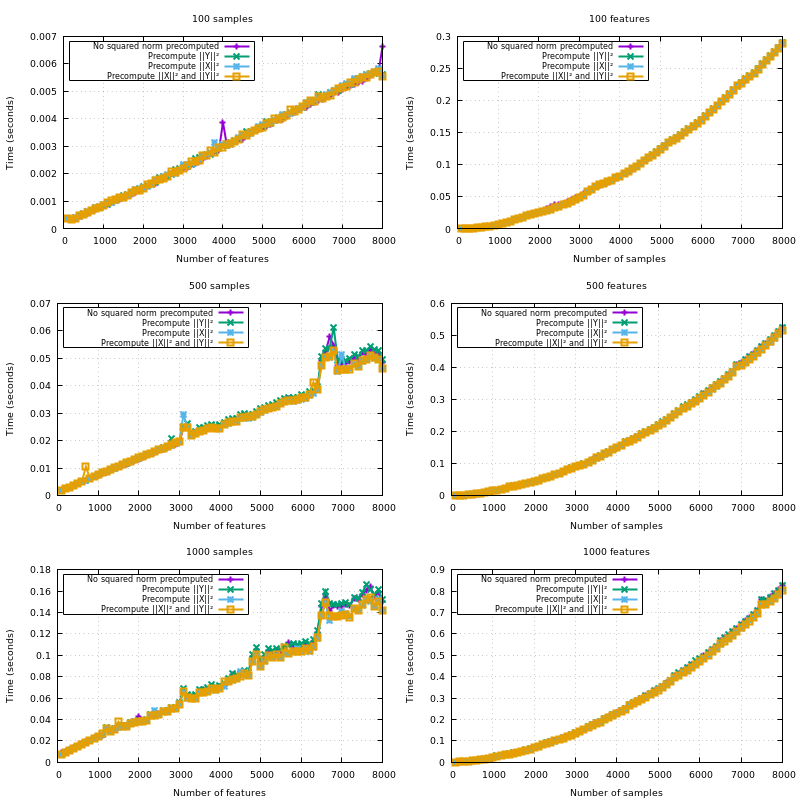
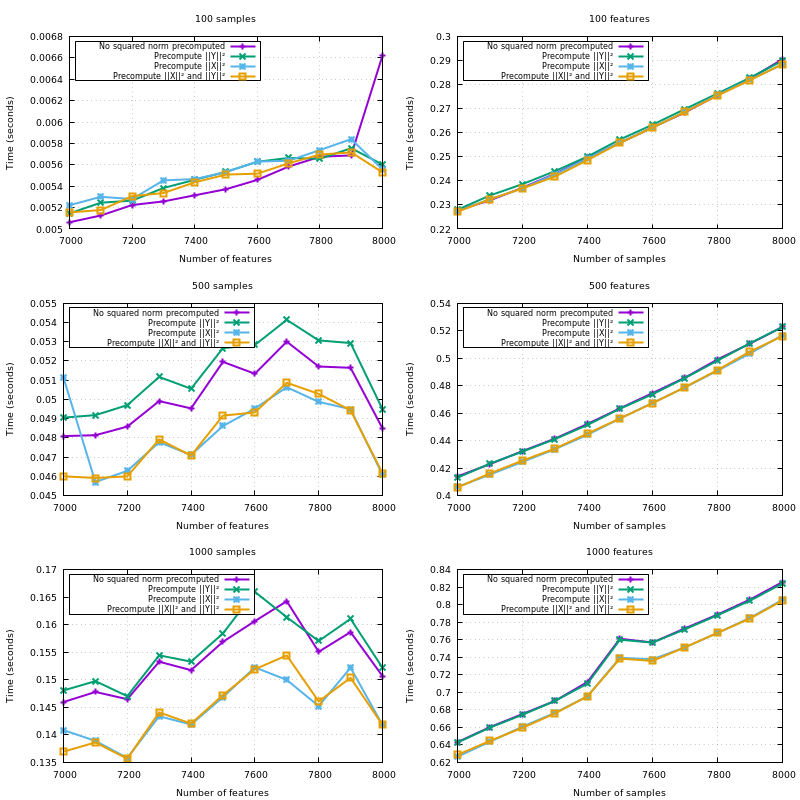
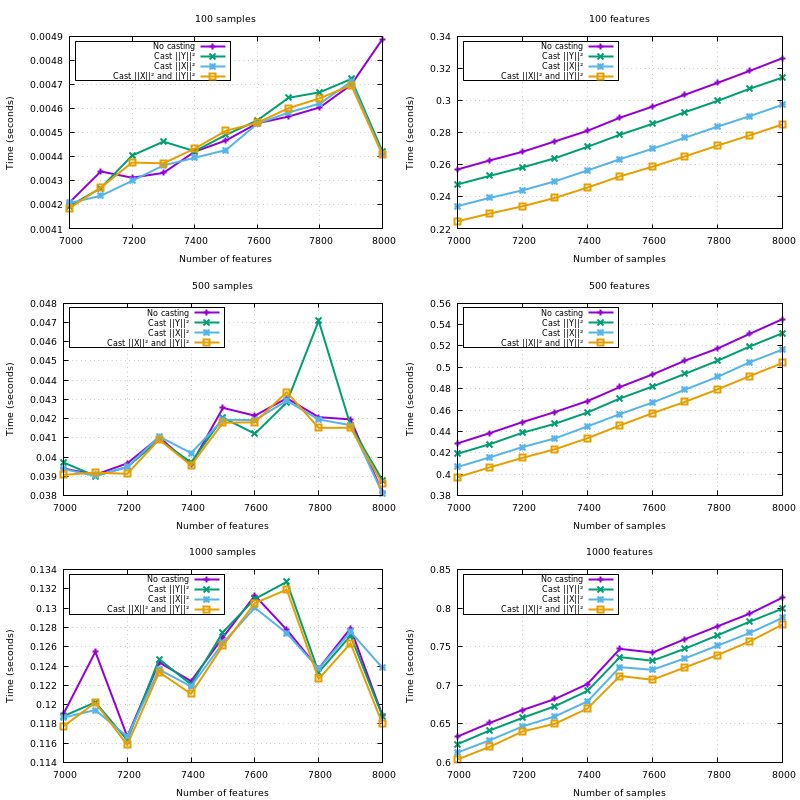
X is Y, copy the blocks of the upper triangle to the lower triangle;{X,Y}_norm_squaredto float64;X_norm_squaredif not given so it gets reused through the iterations overY;XandYwhenX_norm_squaredis given, but notY_norm_squared.Note that all the optimizations listed here have proven useful in a benchmark. The hardcoded amount of additional memory of 10MB is also derived from a benchmark.
As a side bonus, this implementation should also support float16 out of the box, should scikit-learn support it at some point.