OrthoFinder v2.4.0
New in this release
Phylogenetically inferred orthogroups: OrthoFinder now creates a new directory that contains orthogroups defined at each level in the species tree. These orthogroups are inferred by examining the gene trees using the same algorithm that OrthoFinder uses to infer orthologs. Because they are inferred by analysing gene trees they are substantially more accurate than any other method available (and give an approximately 10% relative increase in accuracy on the Orthobench benchmarks compared to OrthoFinder version 2). These files are in the new results directory Phylogenetic_Hierarchical_Orthogroups/.
Because OrthoFinder now infers orthogroups at each phylogenetic level within the species tree it is now possible to include outgroup species in your analysis. Then, to see the orthogroups for just your species of interest just use the corresponding file from the Phylogenetic_Hierarchical_Orthogroups/. The clade names N1, N2, etc. can be found in Species_Tree/SpeciesTree_rooted_node_labels.txt. The use of outgroup species can further increase accuracy (~13% relative increase compared to OrthoFinder v2).
Hierarchical orthogroups are useful because, due to gene duplication events, orthogroups become more fine grained as the species become more closely related:
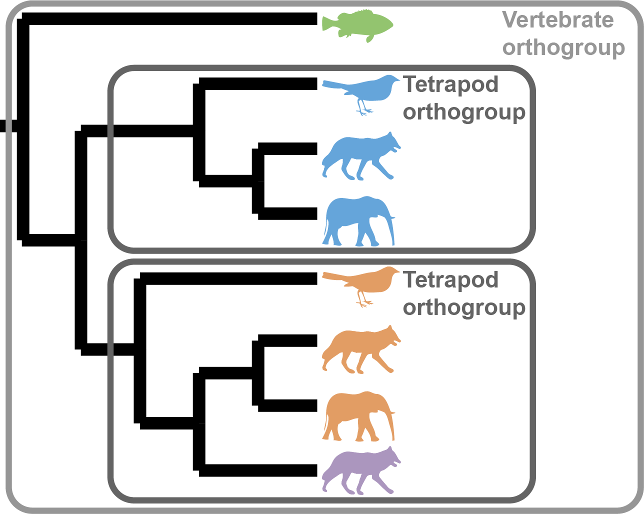
This is the first of a two part series of developments to increase OrthoFinder orthogroup accuracy using the analysis of gene trees.
Which package to download:
- On Linux download OrthoFinder.tar.gz. This bundles all the required external dependencies (mcl, diamond, fastme) and python libraries and so should run immediately, without any installation being required.
- On Mac the bioconda package is probably the easiest method: See Bioconda getting started and, once bioconda is set up, run
conda install orthofinder - On either platform you can run the source code version but you will need to have python and the numpy & scipy libraries installed.
- On Windows the best way is to install the Windows Subsystem for Linux and then use the linux version
More detailed instructions here: https://davidemms.github.io/orthofinder_tutorials/alternative-ways-of-getting-OrthoFinder.html