Evident is a tool for performing effect size and power calculations on microbiome data.
You can install the most up-to-date version of Evident from PyPi using the following command:
pip install evident
Evident is also available as a QIIME 2 plugin. Make sure you have activated a QIIME 2 environment and run the same installation command as above.
To check that Evident installed correctly, run the following from the command line:
qiime evident --help
You should see something like this if Evident installed correctly:
Usage: qiime evident [OPTIONS] COMMAND [ARGS]...
Description: Perform power analysis on microbiome data. Supports
calculation of effect size given metadata covariates and supporting
visualizations.
Plugin website: https://github.com/biocore/evident
Getting user support: Please post to the QIIME 2 forum for help with this
plugin: https://forum.qiime2.org
Options:
--version Show the version and exit.
--example-data PATH Write example data and exit.
--citations Show citations and exit.
--help Show this message and exit.
Commands:
multivariate-effect-size-by-category
Multivariate data effect size by category.
multivariate-power-analysis Multivariate data power analysis.
plot-power-curve Plot power curve.
univariate-effect-size-by-category
Univariate data effect size by category.
univariate-power-analysis Univariate data power analysis.
univariate-power-analysis-repeated-measures
Univariate data power analysis for repeated
measures.
visualize-results Tabulate evident results.
Evident can operate on two types of data:
- Univariate (vector)
- Multivariate (distance matrix)
Univariate data can be alpha diversity. log ratios, PCoA coordinates, etc. Multivariate data is usually a beta diversity distance matrix.
For this tutorial we will be using alpha diversity values, but the commands are nearly the same for beta diversity distance matrices.
First, open Python and import Evident
import evidentNext, load your diversity file and sample metadata.
import pandas as pd
metadata = pd.read_table("data/metadata.tsv", sep="\t", index_col=0)
faith_pd = metadata["faith_pd"]The main data structure in Evident is the 'DataHandler'.
This is the way that Evident stores the data and metadata for power calculations.
For our alpha diversity example, we'll load the UnivariateDataHandler class from Evident.
UnivariateDataHandler takes as input the pandas Series with the diversity values and the pandas DataFrame containing the sample metadata.
By default, Evident will only consider metadata columns with, at max, 5 levels.
We choose 5 samples as the default value because columns with more than 5 groups may be indicative of technical columns (e.g. subject ID).
To modify this behavior, provide a value for the max_levels_per_category argument.
You can set this value to -1 to not provide an upper limit of levels at which to drop a column.
Additionally, Evident will not consider any category levels represented by fewer than 3 samples.
We choose 3 samples as the default value because levels with fewer than 3 samples may exhibit high variances.
To modify this behavior, use the min_count_per_level argument (must be > 1).
adh = evident.UnivariateDataHandler(faith_pd, metadata)Next, let's say we want to get the effect size of the diversity differences between two groups of samples. We have in our example a column in the metadata "classification" comparing two groups of patients with Crohn's disease. First, we'll look at the mean of Faith's PD between these two groups.
metadata.groupby("classification").agg(["count", "mean", "std"])["faith_pd"]which results in
count mean std
classification
B1 99 13.566110 3.455625
Non-B1 121 9.758946 3.874911
Looks like there's a pretty large difference between these two groups. What we would like to do now is calculate the effect size of this difference. Because we are comparing only two groups, we can use Cohen's d. Evident automatically chooses the correct effect size to calculate - either Cohen's d if there are only two categories or Cohen's f if there are more than 2.
adh.calculate_effect_size(column="classification")This tells us that our effect size is 1.03.
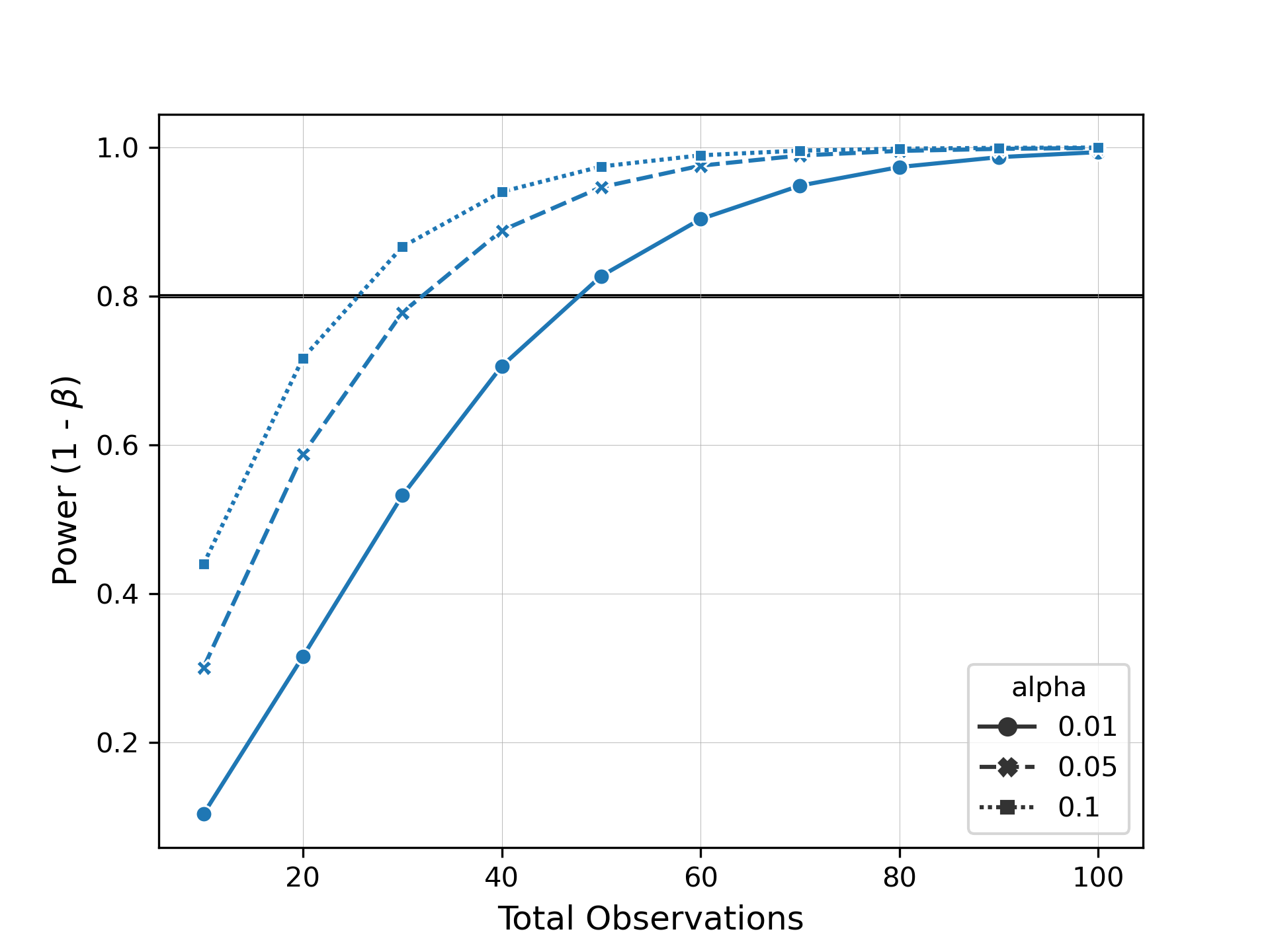
Now let's say we want to see how many samples we need to be able to detect this difference with a power of 0.8. Evident allows you to easily specify arguments for alpha, power, or total observations for power analysis. We can then plot these results as a power curve to summarize the data.
from evident.plotting import plot_power_curve
import numpy as np
alpha_vals = [0.01, 0.05, 0.1]
obs_vals = np.arange(10, 101, step=10)
results = adh.power_analysis(
"classification",
alpha=alpha_vals,
total_observations=obs_vals
)
plot_power_curve(results, target_power=0.8, style="alpha", markers=True)When we inspect this plot, we can see how many samples we would need to collect to observe the same effect size at different levels of significance and power.
Evident allows users to interactively perform effect size and power calculations using Bokeh. To create a Bokeh app, use the following command:
from evident.interactive import create_bokeh_app
create_bokeh_app(adh, "app")This will save the necessary files into a new directory app/.
Navigate to the directory containing app/ (not app/ itself) and execute this command from your terminal:
bokeh serve --show app
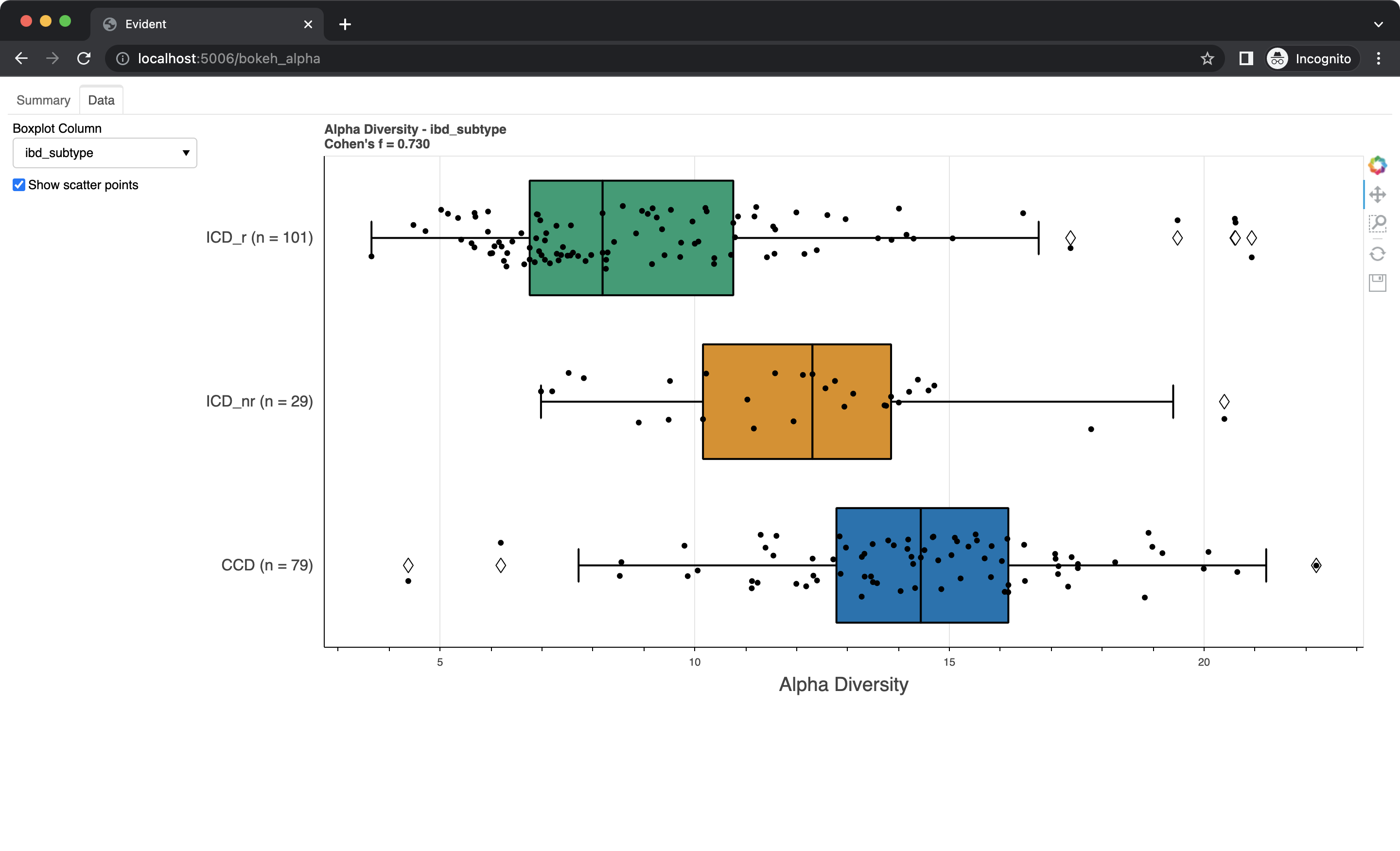
This should open up a browser window with the interactive visualizations. The "Summary" tab gives an overview of the data and the effect sizes/power. Barplots showing the metadata effect sizes for both binary and multi-class categories (ranked in descending order) are shown. On the right is a dynamic power curve showing the power analysis for metadata columns. The significance level, total observation range, and chosen columns can be modified by using the control panel on the left side of the tab.
Swap to the "Data" tab using the bar on the top. Here you can see boxplots of the data for each metadata category. Select a column from the dropdown to change which data is shown. You can also check the "Show scatter points" box to overlay the raw data onto the boxplots.
Note that because evident uses Python to perform the power calculations, it is at the moment not possible to embed this interactive app into a standalone webpage.
Evident provides support for the popular QIIME 2 framework of microbiome data analysis. We assume in this tutorial that you are familiar with using QIIME 2 on the command line. If not, we recommend you read the excellent documentation before you get started with Evident. Note that we have only tested Evident on QIIME 2 version 2021.11. If you are using a different version and encounter an error please let us know via an issue.
To calculate power, we can run the following command:
qiime evident univariate-power-analysis \
--m-sample-metadata-file metadata.qza \
--m-sample-metadata-file faith_pd.qza \
--p-data-column faith_pd \
--p-group-column classification \
--p-alpha 0.01 0.05 0.1 \
--p-total-observations $(seq 10 10 100) \
--o-power-analysis-results results.qza
We provide multiple sample metadata files to QIIME 2 because they are internally merged.
You should provide a value for --p-data-column so Evident knows which column in the merged metadata contains the numeric values (this is only necessary for univariate analysis).
In this case, the name of the faith_pd.qza vector is faith_pd so we use that as input.
Notice how we used $(seq 10 10 100) to provide input into the --p-total-observations argument.
seq is a command on UNIX-like systems that generates a sequence of numbers.
In our example, we used seq to generate the values from 10 to 100 in intervals of 10 (10, 20, ..., 100).
With this results artifact, we can visualize the power curve to get a sense of how power varies with number of observations and significance level. Run the following command:
qiime evident plot-power-curve \
--i-power-analysis-results results.qza \
--p-target-power 0.8 \
--p-style alpha \
--o-visualization power_curve.qzv
You can view this visualization at view.qiime2.org directly in your browser.
Evident provides support for parallelizing effect size calculations through joblib.
Parallelization is performed across different columns when using effect_size_by_category and pairwise_effect_size_by_category.
Consider parallelization if you have a lot of samples and/or a lot of different metadata categories of interest.
By default, no parallelization is performed.
With Python:
from evident.effect_size import effect_size_by_category
effect_size_by_category(
adh,
["classification", "cd_resection", "cd_behavior"],
n_jobs=2
)With QIIME 2:
qiime evident univariate-effect-size-by-category \
--m-sample-metadata-file metadata.qza \
--m-sample-metadata-file faith_pd.qza \
--p-data-column faith_pd \
--p-group-columns classification sex cd_behavior \
--p-n-jobs 2 \
--o-effect-size-results alpha_effect_sizes.qza
If you encounter a bug in Evident, please post a GitHub issue and we will get to it as soon as we can. We welcome any ideas or documentation updates/fixes so please submit an issue and/or a pull request if you have thoughts on making Evident better.
If your question is regarding the QIIME 2 version of Evident, consider posting to the QIIME 2 forum. You can open an issue on the Community Plugin Support board and tag @gibsramen if required.
If you use Evident in your work, please cite our preprint on bioRxiv.