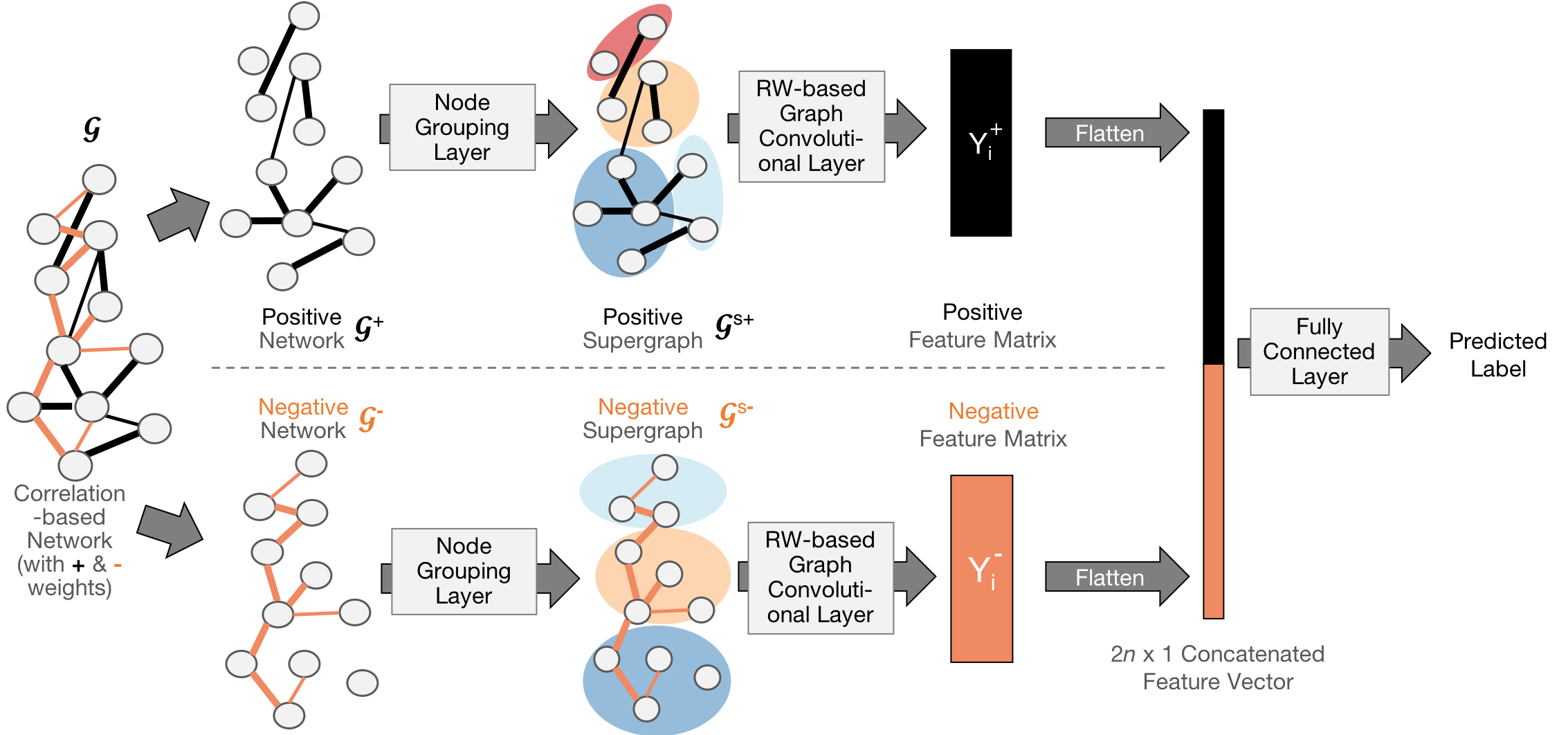
Paper: Yujun Yan, Jiong Zhu, Marlena Duda, Eric Solarz, Chandra Sripada, Danai Koutra. GroupINN: Grouping-based Interpretable Neural Network-based Classification of Limited, Noisy Brain Data. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD), 2019.
Link: https://gemslab.github.io/papers/yan-2019-groupinn.pdf
Citation (bibtex):
@inproceedings{DBLP:conf/kdd/YanZDSSK19,
author = {Yujun Yan and
Jiong Zhu and
Marlena Duda and
Eric Solarz and
Chandra Sripada and
Danai Koutra},
title = {GroupINN: Grouping-based Interpretable Neural Network-based Classification of Limited, Noisy Brain Data},
booktitle = {Proceedings of the 25th {ACM} {SIGKDD} International Conference on
Knowledge Discovery {\&} Data Mining, {KDD} 2019, London, UK,
August 4-8, 2019},
year = {2019},
}
- Python 3.6 and up
- TensorFlow 1.14.0
Due to confidentiality reasons, we are not allowed to publish the Human Connectome data here. However, the Human Connectome data has open access; you can apply to access it through the link.
We are using the Human Connectome Project 1200 release; our collaborators in neuroscience help process the data. The methods to process the data is described in their paper.
The input to our model is time series; data should be in .mat files. Subjects’ data is saved in separate mat files and each subject have multiple mat flies for measurements at different time. The matrix in the mat file has the shape num_timestamps × num_rois. Our code will take care of the graph generation. If you already have graph data at hand, you need to modify foundations/data_manager.py.
The names of brain regions can be found here
train_model.py: main script to train the model and obtain interpretable results on the best checkpoint obtained through training.interpret_model.py: main script for interpretability analysis.models: folder containing model files.- Our model is in
GroupINN.py. - You may modify our model and put it as another python module in this folder, and use
--model_fileargument to run.
- Our model is in
foundations/data_manager.py: data input and preprocessing.foundations/arguments.py: arguments and functional hooks management.foundations/run_training.py: files containing training details.foundations/logging.py: logging and checkpoint management.
To train our model and get the interpretable results on the best checkpoint obtained through training, run
python train_model.py
Two optional arguments which may be of the most interest:
--selected_timeseries [SELECTED_TIMESERIES]: Select timeseries that you would like to feed into your network. Choose from ‘wmMEAN’, ‘emotionMEAN’, ‘gamblingMEAN’, ‘socialMEAN’ (default: 'wmMEAN')--train_epoch [TRAIN_EPOCH]: Total number of epochs to run in the training process (default: 300)
Run python train_model.py -- help to check all available arguments.
After training process, the interpretations will be saved at within_regions.txt and across_regions.txt.
- The
within_regions.txtcontains the sorted most important brain regions; - The
across_regions.txtcontains the sorted most important region pairs.
We saved the best checkpoint path in the variable post_train_dict["best_checkpoint_path"] and printed it out.
If you want to check the statistics of that checkpoint, you can find the best checkpoint path. The way the file is named reflects the statistics:
{Training Acc}-{Validation Acc}-{Test Acc}-{Validation True Positive Rate}-{Validation True Negative Rate}
Email yujunyan@umich.edu or jiongzhu@umich.edu. If your question is more related to acquisition or preprocessing of fMRI time series, you may also want to email csripada@gmail.com for faster response.