-
Notifications
You must be signed in to change notification settings - Fork 8
Home
This wiki constitutes the primary documentation available for the MASTER stochastic simulation system, which can be obtained from the project webpage at http://tgvaughan.github.io/MASTER.
MASTER (Moments and Stochastic Trees from Event Reactions) is a system for generating simulated dynamics under a certain type of stochastic model commonly used to described the dynamics of discrete populations. These models come under the broad heading of continuous time discrete state Markovian models. Specifically, MASTER can simulate the dynamics of processes which can be described in terms of a particular kind of birth-death master equation known as the Chemical (or Combinatoric) Master Equation (CME). Importantly, trees and networks can be simultaneously simulated under the same models. Simulations are specified in a human-readable XML syntax and can be performed using a variety of simulation algorithms. The results of simulations are stored in highly portable JSON, Newick and NEXUS formats.
MASTER is fundamentally a "package" for the BEAST 2 open source phylogenetic inference package. For ease of use, however, it is also distributed in a stand-alone form.
In deciding whether to use MASTER for a particular problem one should consider the goals that it sets out to achieve, which are as follows.
- To be a versatile simulator for the study of continuous time stochastic population dynamics models and trees/networks generated under these models.
- To enable easy distribution of simulation specifications in an easy-to-read format.
- To be extensible: it should be possible to add new simulation algorithms relatively easily.
- To be correct: extensive and ongoing testing allows us to ensure that the implementation is correct.
- To be as computationally efficient as possible, but only as far as the previous goals can still be met.
The question of whether to use MASTER then boils down to a question of how well the goals above align with your own. Generally speaking, if you need to quickly assemble simulations of population sizes and/or genealogies from a variety of different continuous-time stochastic population dynamics models and want to retain the ability to easily modify the model structure, you may find MASTER useful.
On the other hand, if your goal is to perform simulations under a single model and computational efficiency is the primary concern, it may instead make sense to produce your own tailored simulator. Even in that case, however, MASTER may still prove useful in the initial exploratory phase.
Instructions for obtaining and installing MASTER are available on the project webpage at http://tgvaughan.github.com/MASTER.
MASTER functions by taking a file containing an XML description of a stochastic model, performing the necessary calculations and writing the results out to disk in formats depending on the specific type of simulation requested. This process is outlined as follows.

The following sections introduce in more detail the kinds of models that MASTER can handle, the nomenclature that is needed to specify these models within MASTER's framework, and entry points to the full description of the MASTER input file syntax.
As mentioned above, the models which MASTER deals with are those stochastic population dynamics models which can be expressed in terms of the Chemical Master Equation (CME). The CME in turn determines the dynamics of a probability distribution over the possible occupancy states occupied by a system composed of distinct, internally well-mixed populations, when those populations are evolving under the influence of a fixed number of stochastic birth/death processes or "reactions".
The CME itself takes the form
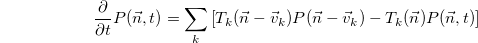
where
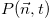 is the probability that the sizes of the populations constituting the
system are equal to the elements of
is the probability that the sizes of the populations constituting the
system are equal to the elements of
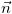 at time t, and
at time t, and
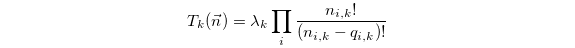
is the propensity with which a reaction k occurs. The variables
 ,
, 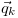 and
and
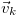 are the reaction rate constant, the reactant
vector and the reaction state transition vector. The rate constant
fixes the rate at which a reaction contributes to the dynamics per
available reactant permutation. The reactant vector specifies the
number of individuals contributing to the reaction from each
population, and the state transition vector determines the effect that
a single reaction event will have on the system state: i.e.,
are the reaction rate constant, the reactant
vector and the reaction state transition vector. The rate constant
fixes the rate at which a reaction contributes to the dynamics per
available reactant permutation. The reactant vector specifies the
number of individuals contributing to the reaction from each
population, and the state transition vector determines the effect that
a single reaction event will have on the system state: i.e.,
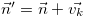 immediately following a reaction of type k.
Together, these variables completely specify a reaction. They can be
expressed succinctly using the notation of chemical kinetics:
immediately following a reaction of type k.
Together, these variables completely specify a reaction. They can be
expressed succinctly using the notation of chemical kinetics:
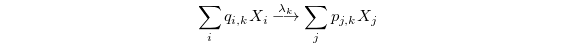
with the understanding that 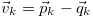 . This basic
form is used to describe all stochastic models that MASTER handles,
and the first step in performing such a simulation is always the
expression of a model in this form.
. This basic
form is used to describe all stochastic models that MASTER handles,
and the first step in performing such a simulation is always the
expression of a model in this form.
In addition to a basic understanding of this model framework, it is important that the user also be familiar with the nomenclature MASTER uses to describe populations, reactions and moments. This is described in the sections below.
In MASTER, a population refers to a group of indistinguishable individuals whose state at a particular time is to be recorded as a single integer specifying the total number of those individuals. At the most basic level, this is all you need to know. However, if you're ever going to be considering models involving more than a very small handful of such populations, please continue reading.
Fundamentally, MASTER deals with groups of populations it refers to as population types. Each type is associated with a unique name. This name can be any single-word string, but by convention is usually a single capital letter. Each population type can be associated with an arbitrary number of actual populations. These can be thought of as sub-populations or demes. MASTER allows these populations to be arranged in an n-dimensional array. A population type is completely specified by both its name and the dimension of the array of populations it contains. The featureless (or "scalar") population referred to in the previous paragraph is simply a population type containing a single deme.
Just as populations are bundled into types, so reactions---the individual components of the stochastic models we consider---are brought together into reaction groups. Each reaction group has a unique name and may contain one or more individual reactions. The point of this is to allow large numbers of very similar reactions such as migrations between individual demes to be logically grouped together so as to make the model specification more readable.
Just as for populations, it is possible to specify a single named reaction.
In some instances, the goal of stochastic simulation is not to produce representative population size histories, but to estimate moments of the population sizes such as the means, variances and covariances. Unlike individual trajectories, these are quantities which can be directly compared with experimental data.
MASTER allows one to estimate the mean and variance of any product of population sizes. Sums of these products can also be estimated. A single product is referred to as a moment. A collection of these can be grouped together into a moment group. Elements of a group can be estimated and recorded independently, or summed together.
This section is the entry point to the wiki pages which describe the details of the XML input format used to specify MASTER simulations. These pages are primarily intended to serve as a reference, so you shouldn't worry too much about reading them in detail until you've completed reading/working through the tutorials. That said, it is important to understand the structure of the input file at the coarsest level.
MASTER inherits its XML format from BEAST 2. Every MASTER XML file must therefore begin with
<beast version='2.0' namespace='master:master.model:master.steppers:master.conditions:master.postprocessors:master.outputs'>and end with the closing element tag </beast>. (Warning: the value of the namespace attribute has changed in versions 1.5 and later.) This outermost element must contain the following element:
<run spec='Trajectory|Ensemble|EnsembleSummary|InheritanceTrajectory|InheritanceEnsemble'>
<!-- Simulation specification -->
</run>The value of the attribute spec determines which type of simulation
to run. There are currently 5 distinct simulation types. While the
overall format of the simulation specification is very similar for
each type, there are differences. The links below are to pages
describing the details of the specification format for each simulation
type.
- Trajectory specification - single population size simulations
- Ensemble specification - multiple population size simulations
- EnsembleSummary specification - multiple population size simulations, with on-the-fly estimation of summary statistics.
- InheritanceTrajectory specification - single simulation including (not limited to) inheritance graph generation
- InheritanceEnsemble specification - multiple inheritance graph simulations
Beyond these principal calculation types, there are two additional types that are of use when combining MASTER simulations with other BEAST analyses:
- BeastTreeFromMaster can be used to initialise BEAST tree objects, which can then be used as the basis for simulating alignments or simply for initialising MCMC runs.
- PopulationFunctionFromMaster allows MASTER trajectories to be used as part of BEAST's coalescent framework.
The tutorials below introduce the major features of MASTER. This should be the starting point for novice users once the software itself has been downloaded and installed, as per the instructions on the main page. They are presented in order of increasing complexity, with later tutorials assuming that the user has read through the previous tutorials.
-
Tutorial 1: Simulating dynamics under a stochastic logistic model. (Basic usage and output processing.)
-
Tutorial 2: Estimating moments from an ensemble of realizations of an island migration model. (Stochastic moment estimation, structured populations, end conditions.)
-
Tutorial 3: Simulating an infection transmission tree from an epidemic model. (Inheritance graph simulation, inheritance graph output.)
-
Tutorial 4: Structured coalescent trees. (Explicit inheritance relationship specification, reverse-time output.)
-
Tutorial 5: One-dimensional random walk. (Location index variables, predicates, rate multipliers, functions.) INCOMPLETE
If you use MASTER as part of research that leads to a publication, we ask that you cite the following article:
T. G. Vaughan and A. J. Drummond, "A Stochastic Simulator of Birth–Death Master Equations with Application to Phylodynamics", Mol. Biol. Evol. 30(6):1480, 2013. (link)

Development of MASTER is generously supported by the Allan Wilson Centre for Molecular Ecology and Evolution.