A Bioinformatics Tool for the Integrative Analysis of Alternative Splicing Regulome using RNA-Seq data across cancer and tissue types
regulAS is a computational software for analysis of regulatory mechanisms of alternative splicing enabling to dissect complex relationships between alternative splicing events and thousands of its prospective regulatory RBPs using large-scale RNA-seq data derived from The Cancer Genome Atlas (TCGA) and The Genotype-Tissue Expression (GTEx) projects.
regulAS is a powerful machine learning experiment management tool written in Python that combines
flexible configuration system based on YAML and shell, fast and reliable data storage
driven by SQLite relational database engine and an extension-friendly API compatible
with scikit-learn.
regulAS is designed to provide users with a "Low-Code" experiment management solution that is dedicated for researchers of alternative splicing regulatory mechanisms to simplify computational workflow, allowing them to alleviate the number of the prospective regulatory candidates of splicing changes for further in-depth bioinformatics and experimental analysis.
regulAS is supplied with a set of pre-defined modules that introduce support for data acquisition, fitting of machine learning models, persistence and export of results.
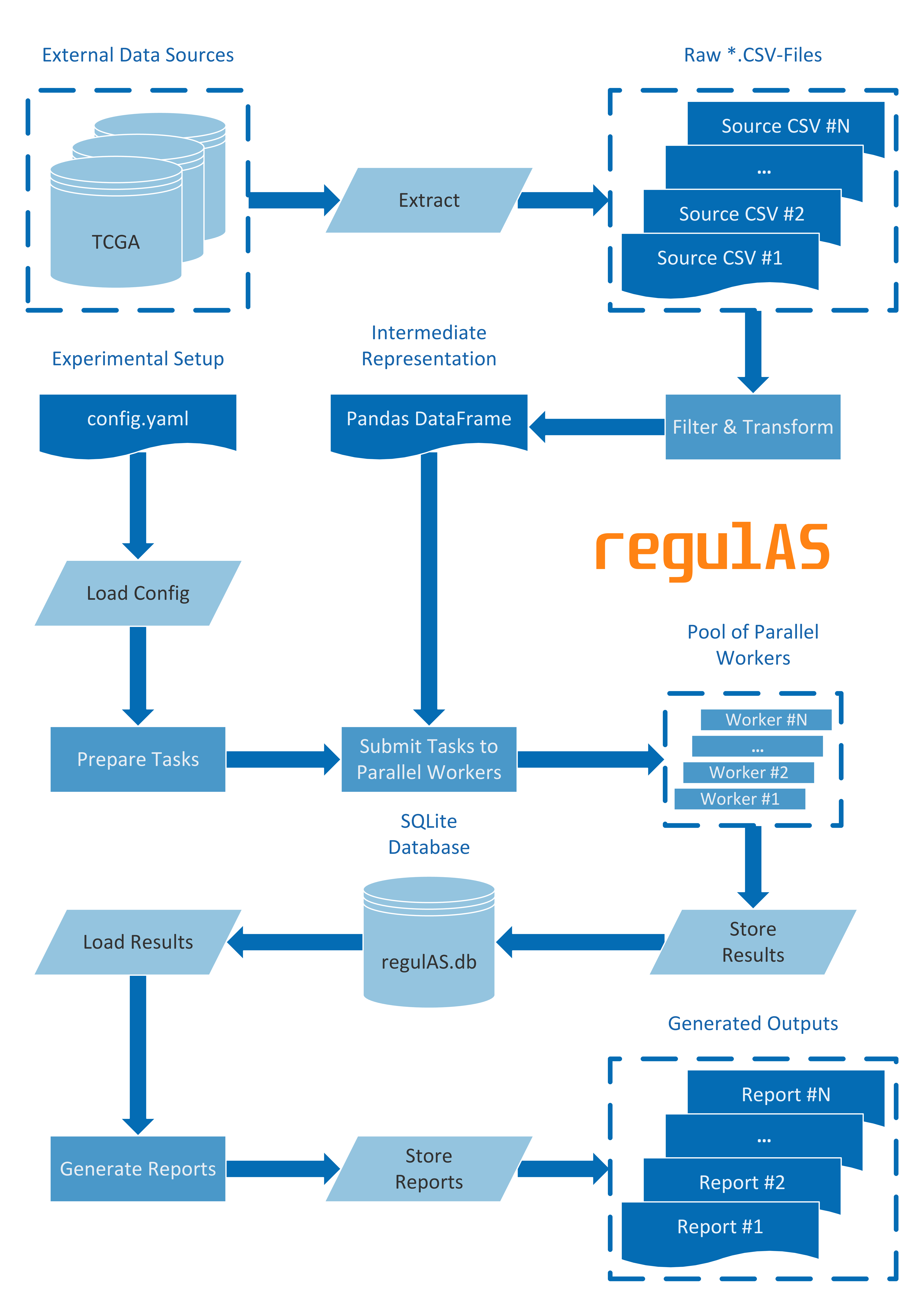
The regulAS package encapsulates ETL-, ML- and report generation workflows. ETL-workflow includes data Extraction, Transformation and Loading operations, thus preparing the input for the next step, namely Machine Learning (ML) part. The ML-workflow incorporates predictive modeling, hyper-parameter optimization, performance evaluation and feature ranking tasks for identifying candidate regulators of alternative splicing events across tumors and tissue types. The ML-workflow outputs can subsequently be utilized for generating summary reports in tabular and visual forms to support interpretation of the findings and knowledge sharing.
After the transformation step, regulAS loads the data into a pandas.DataFrame,
which is a default container to store intermediate results.
Given an additional argument denoting the target for supervised model training,
data loader splits the DataFrame into samples and targets that are used by the ML-part.
Experimental setups are configured through YAML files using Facebook Hydra framework.
The Machine Learning sub-flow manages hyper-parameter tuning, performance evaluation
and scoring of feature relevance.
To tune hyper-parameters, regulAS performs a cross validation procedure that is similar
to the scikit-learn grid search (GridSearchCV).
The model performance evaluation requires comparison of ground truth targets to the actual predictions.
Thus, both true and predicted values are stored into a database that serves as a data source
for performance evaluation reports.
Finally, regulAS detects if a model provides information on feature importance
and intercepts this scoring putting it into a database for later analysis.
regulAS tracks and stores a wide variety of experimental data that includes:
- Dataset description
- Experimental configuration
- Actual hyper-parameters
- Source code for models
- True and predicted target values
- Feature importance
Typically, a single experiment is able to provide the management system with several hundred thousand records, which makes storing them a non-trivial task. To address this challenge, regulAS relies on a relational database powered by the SQLite engine. The use of the SQLite database ensures data integrity and provides fast and reliable storage for it.
The report generation workflow allows assembling results and summarizing them in an appropriate way (e.g., text- or image-based output). Moreover, the reports can be chained such as the output of a preceding report is fed into the next one depending on it.
How to use YAML configuration files and (optionally) combine them with command line arguments.
A single experimental setup defines a scope named experiment that encapsulates
the following sub-scopes:
name– name of the experimentdataset– configuration of a data sourcesplit– cross validation parameterspipelines– list of ML-configurations to comparereports– data export configurations
Using Facebook Hydra framework, regulAS provides the ability to override configuration
keys from the command line, to define required-in-runtime arguments (using reserved value ???)
and to substitute values given a key to another field of a configuration file (${experiment.another_field}).
regulAS configuration can define model evaluation tasks only, while keeping the list of reports empty.
experiment_tasks.yaml
name: lr_svr_tmr
dataset:
_target_: regulAS.utils.PickleLoader
name: RNA-Seq
meta: some fancy description
path_to_file: ???
objective: psi
split:
_target_: sklearn.model_selection.ShuffleSplit
n_splits: 5
test_size: 0.2
train_size: null
random_state: ${random_state}
pipelines:
- transformations:
ZScore:
_target_: sklearn.preprocessing.StandardScaler
model:
LinearRegression:
_target_: sklearn.linear_model.ElasticNet
l1_ratio: 0.2
_varargs_:
alpha: [0.1, 0.5, 1.0]
- transformations:
MinMax:
_target_: sklearn.preprocessing.MinMaxScaler
model:
SupportVectorMachine:
_target_: sklearn.svm.SVR
kernel: linear
_varargs_:
C: [0.1, 0.5, 1.0]
reports: nullSimilarly, there can be only report tasks defined in the experimental setup.
report_tasks.yaml
name: lr_svr_tmr
dataset: null
split: null
pipelines: null
reports:
MSE:
_target_: regulAS.reports.ModelPerformanceReport
experiment_name: ${experiment.name}
loss_fn: sklearn.metrics.mean_squared_error
PearsonR:
_target_: regulAS.reports.ModelPerformanceReport
experiment_name: ${experiment.name}
score_fn: scipy.stats.pearsonr
FeatureRanking:
_depends_on_: ["MSE"]
_target_: regulAS.reports.FeatureRankingReport
experiment_name: ${experiment.name}
sort_by: "${experiment.reports.MSE.loss_fn}:test:mean"
sort_ascending: true
top_k_models: 3
top_k_features: null
PerformanceCSV:
_depends_on_: ["MSE", "PearsonR"]
_target_: regulAS.reports.ExportCSV
output_dir: reports
sep: ";"
RankingCSV:
_depends_on_: ["FeatureRanking"]
_target_: regulAS.reports.ExportCSV
output_dir: reports
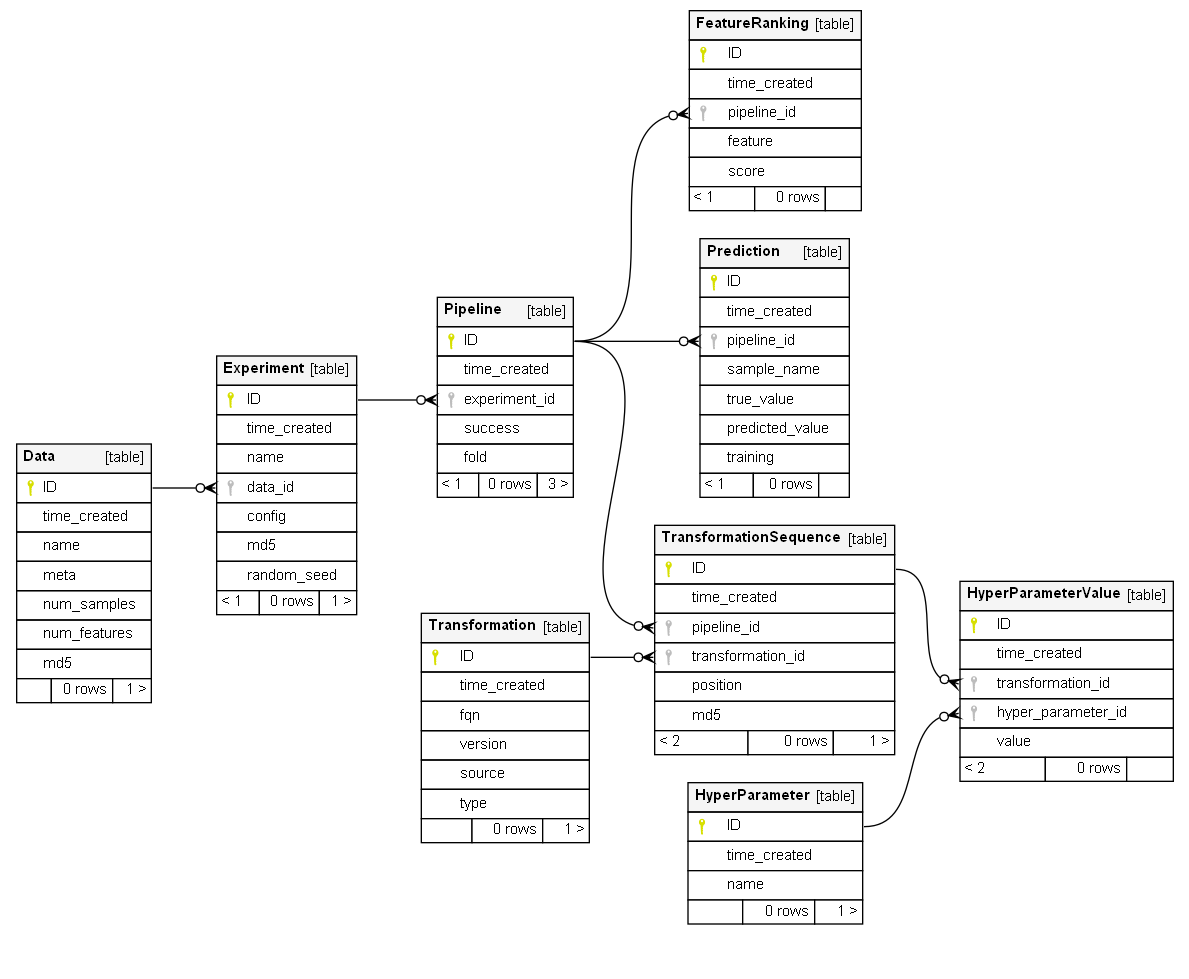
sep: ";"Database structure is dedicated to reliably store experimental results and to serve as a data source for reports based on its contents. Schema of the regulAS' database was designed to preserve consistency and to minimize redundancy of the stored records. The database includes the following tables:
Experiment– contains high-level summary of the experimentsData– contains details on the dataset used inExperimentPipeline– contains high-level summary on specific pipelines used inExperimentTransformation– contains details on ML-models and transformationsHyperParameter– contains hyper-parameters of modelsPrediction– contains true and predicted values for targetFeatureRanking– contains feature importance scoresTransformationSequence– contains details on the use of models and transformationsHyperParameterValue– contains details on the specific values of hyper-parameters
In order to use regulAS for experimental management, user should define a project directory containing YAML configuration files and data if necessary. The experiment then can be run from the command line.
# install regulAS package from PyPi
pip install regulAS
# navigate to the user's project folder
cd /path/to/the/project
# run the experimental setup on different dataset files
python -m regulAS.app --multirun \
experiment=experiments/experiment_tasks \
+dataset.path_to_file=data/data_cNormal.pkl,data/data_cTumor.pklAfter regulAS finishes all the tasks, an SQLite database file regulAS.db will be stored
in the project directory as well as the reports if any was submitted to generate.
When using regulAS in academic work, authors are encouraged to reference this work via Zenodo (10.5281/zenodo.8152781)
An example acknowledgment statement follows: The analysis used for the results described in this manuscript was obtained using the regulAS package.