install aggmap by:
# create an aggmap env
conda create -n aggmap python=3.8
conda activate aggmap
pip install --upgrade pip
pip install aggmap==1.2.1import pandas as pd
from sklearn.datasets import load_breast_cancer
from aggmap import AggMap, AggMapNet
# Data loading
data = load_breast_cancer()
dfx = pd.DataFrame(data.data, columns=data.feature_names)
dfy = pd.get_dummies(pd.Series(data.target))
# AggMap object definition, fitting, and saving
mp = AggMap(dfx, metric = 'correlation')
mp.fit(cluster_channels=5, emb_method = 'umap', verbose=0)
mp.save('agg.mp')
# AggMap visulizations: Hierarchical tree, embeddng scatter and grid
mp.plot_tree()
mp.plot_scatter(enabled_data_labels=True, radius=5)
mp.plot_grid(enabled_data_labels=True)
# Transoformation of 1d vectors to 3D Fmaps (-1, w, h, c) by AggMap
X = mp.batch_transform(dfx.values, n_jobs=4, scale_method = 'minmax')
y = dfy.values
# AggMapNet training, validation, early stopping, and saving
clf = AggMapNet.MultiClassEstimator(epochs=50, gpuid=0)
clf.fit(X, y, X_valid=None, y_valid=None)
clf.save_model('agg.model')
# Model explaination by simply-explainer: global, local
simp_explainer = AggMapNet.simply_explainer(clf, mp)
global_simp_importance = simp_explainer.global_explain(clf.X_, clf.y_)
local_simp_importance = simp_explainer.local_explain(clf.X_[[0]], clf.y_[[0]])
# Model explaination by shapley-explainer: global, local
shap_explainer = AggMapNet.shapley_explainer(clf, mp)
global_shap_importance = shap_explainer.global_explain(clf.X_)
local_shap_importance = shap_explainer.local_explain(clf.X_[[0]])- AggMap flowchart of feature mapping and agglomeration into ordered (spatially correlated) multi-channel feature maps (Fmaps)
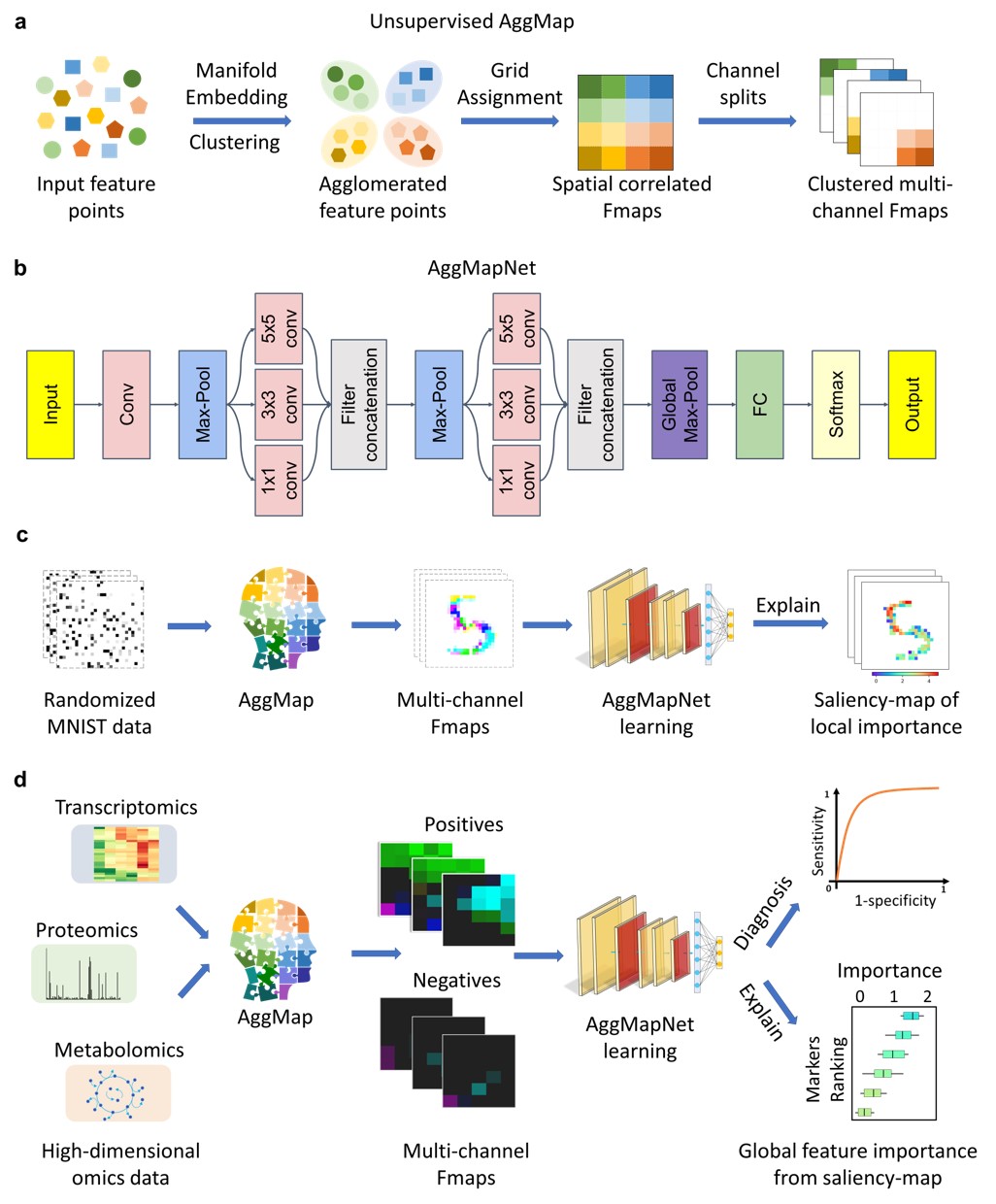 a, AggMap flowchart of feature mapping and aggregation into ordered (spatially-correlated) channel-split feature maps (Fmaps).b, CNN-based AggMapNet architecture for Fmaps learning. c, proof-of-concept illustration of AggMap restructuring of unordered data (randomized MNIST) into clustered channel-split Fmaps (reconstructed MNIST) for CNN-based learning and important feature analysis. d, typical biomedical applications of AggMap in restructuring omics data into channel-split Fmaps for multi-channel CNN-based diagnosis and biomarker discovery (explanation
a, AggMap flowchart of feature mapping and aggregation into ordered (spatially-correlated) channel-split feature maps (Fmaps).b, CNN-based AggMapNet architecture for Fmaps learning. c, proof-of-concept illustration of AggMap restructuring of unordered data (randomized MNIST) into clustered channel-split Fmaps (reconstructed MNIST) for CNN-based learning and important feature analysis. d, typical biomedical applications of AggMap in restructuring omics data into channel-split Fmaps for multi-channel CNN-based diagnosis and biomarker discovery (explanation saliency-map of important features).
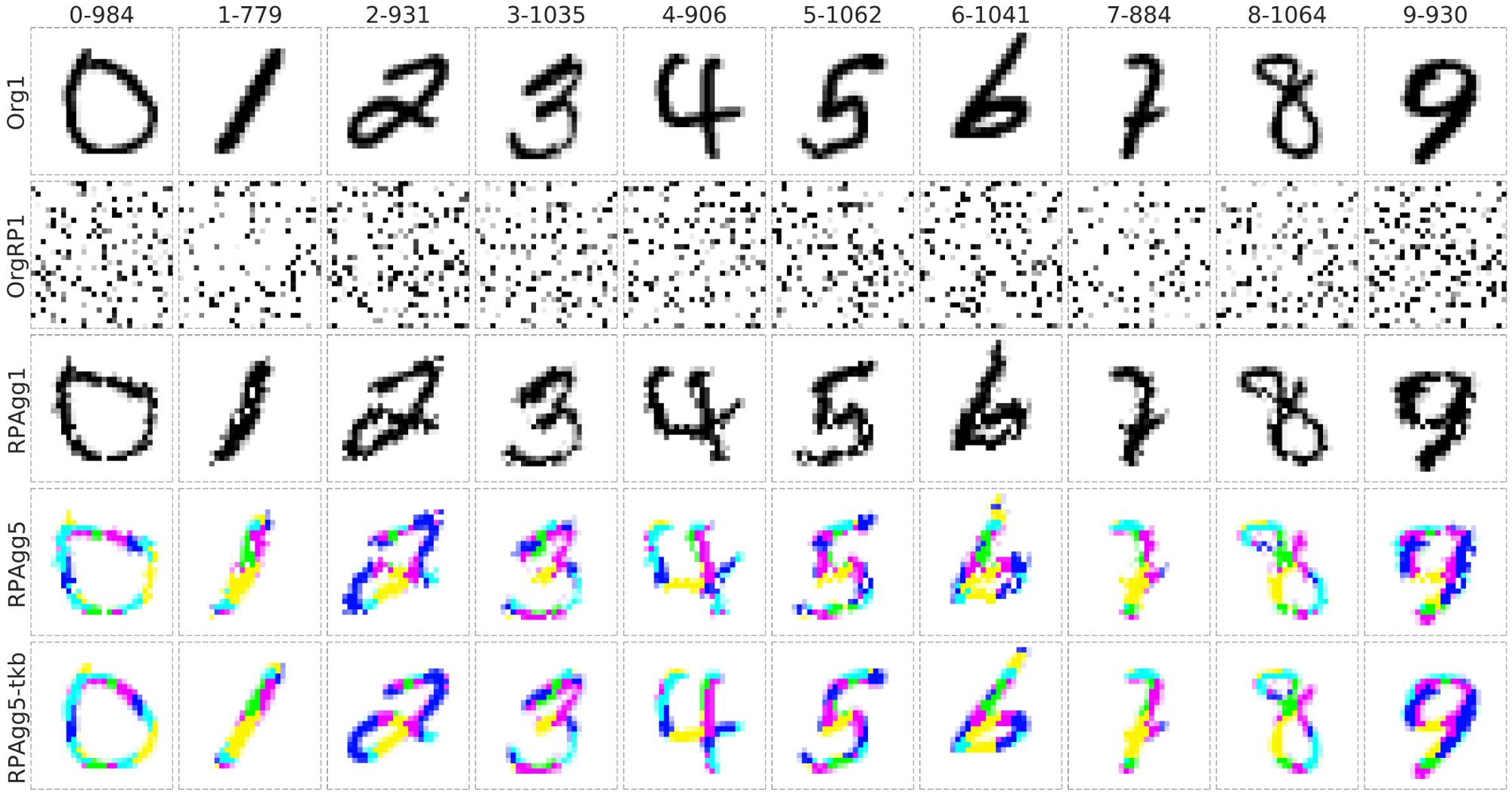
- It can reconstruct to the original image from completely randomly permuted (disrupted) MNIST data:
Org1: the original grayscale images (channel = 1), OrgRP1: the randomized images of Org1 (channel = 1), RPAgg1, 5: the reconstructed images of OrgPR1 by AggMap feature restructuring (channel = 1, 5 respectively, each color represents features of one channel). RPAgg5-tkb: the original images with the pixels divided into 5 groups according to the 5-channels of RPAgg5 and colored in the same way as RPAgg5.
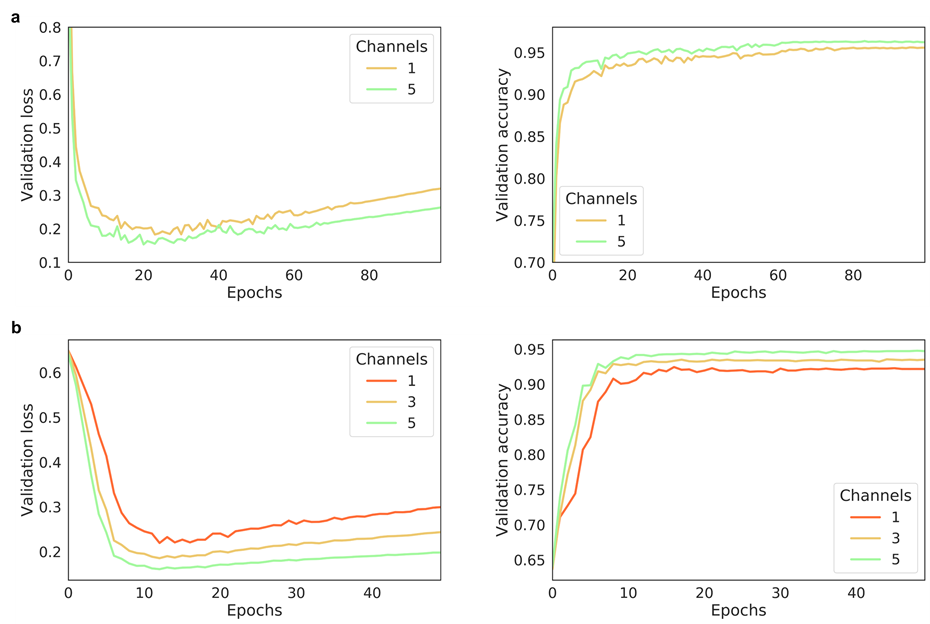
The performance of AggMapNet using different number of channels on the TCGA-T (a) and COV-D (b). For TCGA-T, ten-fold cross validation average performance, for COV-D, a fivefold cross validation was performed and repeat 5 rounds using different random seeds (total 25 training times), their average performances of the validation set were reported.
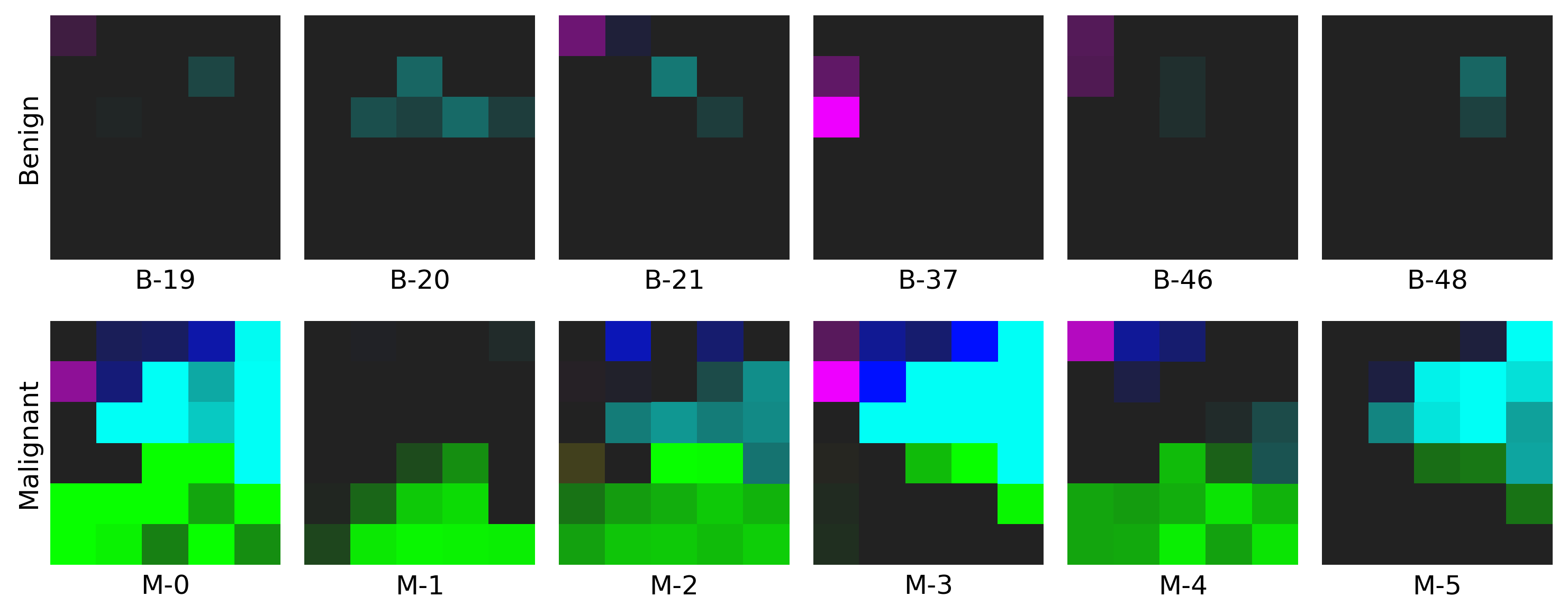
- The example on WDBC dataset: click here to find out more!
Shen, Wan Xiang, et al. "AggMapNet: enhanced and explainable low-sample omics deep learning with feature-aggregated multi-channel networks." Nucleic Acids Research 50.8 (2022): e45-e45.