A Julia package implementing several topic models used for mutation signature estimation.
The Multi-modal correlated topic model (MMCTM) takes an array of arrays of matrices, where the first column of each matrix is a mutation type index, and the second column is the mutation count for a particular sample.
The following example shows how to perform inference using the MMCTM on SNV and SV counts:
using MultiModalMuSig
using CSV
using DataFrames
using VegaLite
using Random
Random.seed!(42)
snv_counts = CSV.read("data/brca-eu_snv_counts.tsv", DataFrame, delim='\t')
sv_counts = CSV.read("data/brca-eu_sv_counts.tsv", DataFrame, delim='\t')
samples = [c for c in propertynames(snv_counts) if c != :term] # names of columns with counts
X = format_counts_mmctm([snv_counts, sv_counts], samples)
model = MMCTM([7, 7], [0.1, 0.1], X)
fit!(model, tol=1e-5)
snv_signatures = DataFrame(hcat(model.ϕ[1]...), :auto)
sv_signatures = DataFrame(hcat(model.ϕ[2]...), :auto)
snv_signatures[!, :term] = snv_counts[!, :term]
snv_signatures = stack(
snv_signatures, Not(:term), variable_name=:signature, value_name=:probability
)
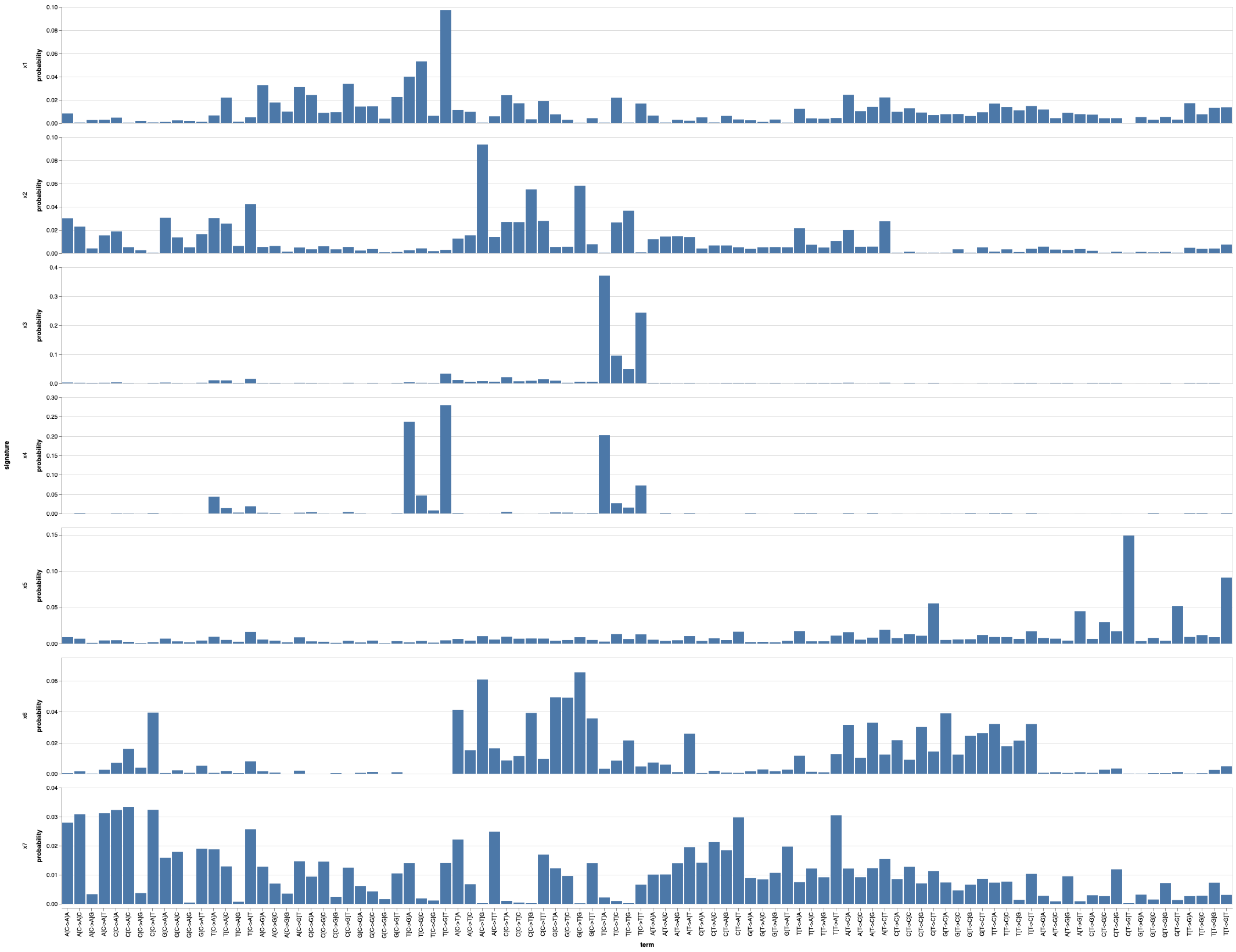
snv_signatures |> @vlplot(
:bar, x={:term, sort=:null}, y=:probability, row=:signature,
resolve={scale={y=:independent}}
)This code runs the MMCTM for 7 SNV and 7 SV signatures, with signature hyperparameters set to 0.1. Since these types of models can get stuck in poor local optima, it's a good idea to fit many models and pick the best one.
To find the set of COSMIC signatures that best match your inferred set of signatures, one approach could be to first compute the cosine distance between the inferred and COSMIC signatures. Then you could use a linear sum assignment solver to find the optimal set of unique matches.
Sample-signature probabilities can be extracted like so:
# sample 3, SNV signature probabilities (modality 1)
model.props[3][1]
# SV signature probabilities (modality 2)
model.props[3][2]
# SNV probabilities for all samples
snv_props = hcat(
[model.props[i][1] for i in 1:length(model.props)]...
)The MMCTM can be run on multiple modalities, e.g.
X = format_counts_mmctm([snv_counts, sv_counts, indel_counts], samples)
model = MMCTM([7, 7, 5], [0.1, 0.1, 0.1], X)To run the CTM instead, just run the MMCTM with a single modality:
X = format_counts_ctm(snv_counts, samples)
model = MMCTM([7], [0.1], X)
fit!(model, tol=1e-5)The LDA implementation can be run like so:
X = format_counts_lda(snv_counts, samples)
model = LDA(7, 0.1, 0.1, X)
fit!(model, tol=1e-5)In the above code, both the sample-signature and signature-term hyperparameters have been set to 0.1, respectively. After fitting LDA, signatures can be found in model.β, and signature probabilities can be found in model.θ.