Toolkit Rosetta
John Mayfield edited this page Sep 13, 2017
·
8 revisions
IChemObjectBuilder builder = SilentChemObjectBuilder.getInstance();
boolean skipErrors = true;
try (InputStream in = new FileInputStream(new File("myinput.sdf"));
Reader rdr = new InputStreamReader(in, StandardCharsets.UTF_8);
IteratingSDFReader sdfr = new IteratingSDFReader(rdr,
builder,
skipErrors)) {
while (sdfr.hasNext()) {
IAtomContainer mol = sdfr.next();
int numHeavy = 0;
for (IAtom atom : mol.atoms()) {
if (atom.getAtomicNumber() > 1)
numHeavy++;
}
}
} catch (FileNotFoundException e) {
System.err.println("No such file: " + e.getMessage());
} catch (IOException e) {
System.err.println("Low level IO error: " + e.getMessage());
}try (InputStream in = new FileInputStream(new File("input.smi"));
Reader rdr = new InputStreamReader(in, StandardCharsets.UTF_8);
BufferedReader brdr = new BufferedReader(rdr)) {
IChemObjectBuilder builder = SilentChemObjectBuilder.getInstance();
SmilesParser smipar = new SmilesParser(builder);
String line;
while ((line = brdr.readLine()) != null) {
try {
IAtomContainer mol = smipar.parseSmiles(line);
int numRings = Cycles.markRingAtomsAndBonds(mol);
} catch (InvalidSmilesException ex) {
System.err.println("BAD SMILES: " + line);
}
}
} catch (FileNotFoundException e) {
System.err.println("No such file: " + e.getMessage());
} catch (IOException e) {
System.err.println("Low level IO error: " + e.getMessage());
}try (InputStream in = new FileInputStream(new File("input.smi"));
Reader rdr = new InputStreamReader(in, StandardCharsets.UTF_8);
BufferedReader brdr = new BufferedReader(rdr)) {
IChemObjectBuilder builder = SilentChemObjectBuilder.getInstance();
SmilesParser smipar = new SmilesParser(builder);
SmilesGenerator smigen = new SmilesGenerator(SmiFlavor.Canonical | SmiFlavor.Stereo);
// N.b. many flavor options to choose
String line;
while ((line = brdr.readLine()) != null) {
try {
IAtomContainer mol = smipar.parseSmiles(line);
System.out.println(smigen.create(mol));
} catch (InvalidSmilesException ex) {
System.err.println("BAD SMILES: " + line);
}
}
} catch (FileNotFoundException e) {
System.err.println("No such file: " + e.getMessage());
} catch (IOException e) {
System.err.println("Low level IO error: " + e.getMessage());
}IChemObjectBuilder builder = SilentChemObjectBuilder.getInstance();
boolean skipErrors = true;
try (InputStream in = new FileInputStream(new File("myinput.sdf"));
Reader rdr = new InputStreamReader(in, StandardCharsets.UTF_8);
IteratingSDFReader sdfr = new IteratingSDFReader(rdr,
builder,
skipErrors)) {
double min = 300;
double max = 400;
int count = 0;
while (sdfr.hasNext()) {
IAtomContainer mol = sdfr.next();
double weight = AtomContainerManipulator.getMolecularWeight(mol);
if (weight >= min && weight <= max)
count++;
}
} catch (FileNotFoundException e) {
System.err.println("No such file: " + e.getMessage());
} catch (IOException e) {
System.err.println("Low level IO error: " + e.getMessage());
}IChemObjectBuilder bldr = SilentChemObjectBuilder.getInstance();
SmilesParser smipar = new SmilesParser(bldr);
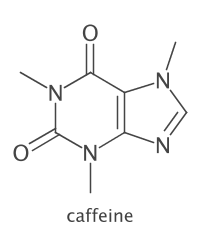
IAtomContainer mol = smipar.parseSmiles("CN1C=NC2=C1C(=O)N(C(=O)N2C)C caffeine");
mol.setProperty(CDKConstants.TITLE, "caffeine"); // title already set from input!
DepictionGenerator dptgen = new DepictionGenerator();
dptgen.withSize(200, 250) // px (raster) or mm (vector)
.withMolTitle()
.withTitleColor(Color.DARK_GRAY) // annotations are red by default
.depict(mol)
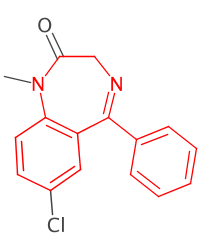
.writeTo("~/caffeine.png"); // svg preferred but will be 20cmx25cm!String fname = "benzodiazepine.sdf";
String sma = "c1ccc2c(c1)C(=NCCN2)c3ccccc3";
IChemObjectBuilder bldr = SilentChemObjectBuilder.getInstance();
IAtomContainer mol = null;
try (FileInputStream in = new FileInputStream(fname);
MDLV2000Reader mdlr = new MDLV2000Reader(in)) {
while ((mol = mdlr.read(bldr.newInstance(IAtomContainer.class))) != null)
if (mol.getProperty(CDKConstants.TITLE).equals("3016"))
break;
}
AtomContainerManipulator.suppressHydrogens(mol);
Pattern ptrn = SmartsPattern.create(sma, bldr);
DepictionGenerator dptgen = new DepictionGenerator();
dptgen.withSize(200, 250)
.withHighlight(ptrn.matchAll(mol)
.uniqueAtoms()
.toChemObjects(),
Color.RED)
.depict(mol)
.writeTo("~/3016.png");