Standard Generator
The standard generator aims to generate aesthetically pleasing depictions that most would hopefully consider acceptable for publication. This page provides technical details, documentation, and showcases examples of using and configuring the generator. The implementation is based heavily on ideas from Brecher (2008) and Clark (2013).
The CDK generators take a structure representation with assigned coordinates and produce the primitive shapes needed to draw a depiction. The process is distinct from structure diagram generation that aims to position or layout the atoms in 2D. The input to the standard generate is therefore an already laid out diagram.
Unlike other generators (such as the BasicAtomGenerator and BasicBondGenerator) atoms and bonds are generated by a single generator instance. This allows the bond generation to know about and avoid the space occupied by each atom symbol. For clarity this document divides up the atom and bond generation.
To use the standard generator, simply supply it to the renderer with the BasicSceneGenerator (required for scaling). The font must be provided at construction. The font used in the depictions on this page was Verdana 18 pt.
Font font = new Font("Verdana", Font.PLAIN, 18);
// create the renderer, note the AWTFontManger isn't used but
// is provided to avoid NPE
AtomContainerRenderer renderer
= new AtomContainerRenderer(Arrays.asList(new BasicSceneGenerator(),
new StandardGenerator(font),
new AWTFontManager()); An example of using a different font can be seen on the Standard Generator Comparison page.
The depictions on this page are rendered as PNG and subject to in interpolation. For best results structures should be rendered as vector graphics (SVG, EPS, and PDF). The Standard Generator Samples page shows some generated SVG samples of some ChEBI structures.
The visibility of atoms is set with the rendering parameter StandardGenerator.Visibility. The parameter takes a SymbolVisibility instance that can have a custom implementation. There are several defaults that can be accessed by a static method on the class.



Left: SymbolVisibility.iupacRecommendations()
Middle: SymbolVisibility.iupacRecommendationsWithoutTerminalCarbon() (default)
Right: SymbolVisibility.all()
The parameter is set on the render model.
RendererModel rendererModel = renderer.getRenderer2DModel();
rendererModel.set(StandardGenerator.Visibility.class,
SymbolVisibility.iupacRecommendations());It is also beneficial to display symbols for emphasis. This is covered in the Emphasis depiction section.
The atom color is controlled by an IAtomColorer instance. This can be set with the StandardGenerator.AtomColor parameter. The bond color is determined using the carbon color.
The default colouring of atom symbols is uniform off black (dark grey). This is suitable for computational display but pitch black is preferable for printed material. This can be set as follows.
rendererModel.set(StandardGenerator.AtomColor.class,
new UniColor(Color.BLACK));Other colors can also be used:
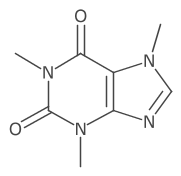
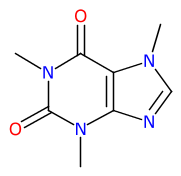
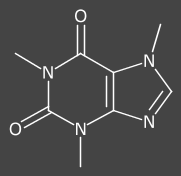
Left: new UniColor(new Color(0x444444)); (default)
Middle: new CDK2DAtomColors();
Right: new UniColor(Color.WHITE); (dark background drawn)
As discussed by Clark (2013), font rendering is tricky. For raster outputs (PNG, JPG, etc), the diagram will look the same (or close enough) independent of the platform it is viewed. Vector outputs (SVG, EPS, PDF, etc) is more difficult as the font used to generate the depiction may not be available.
The fonts used by the standard generator are then first converted to the glyph information and rendered instead as a shape composed of Bézier curves. This is made clearer if we take a look at an enlarged rendering of water and visualize the path points of each character.
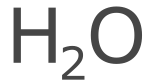
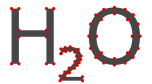
Having access to the text shape allows the precise computation of bond intersects by computing the convex hull of the atom label (using ConvexHull).
The convex hull wraps each atom symbol. How this is used with bond intersects will be discussed further in bond generation.

The text shape in the standard generator is represented as the persistent TextOutline class. Multiple outlines are then composed into the AtomSymbol class that holds information on the element label, hydrogen position, alignment, and zero or more adjuncts. The AtomSymbol also provides access to the ConvexHull.
AtomSymbol {
TextOutline element;
TextOutline[] adjuncts;
ConvexHull hull;
}
A key point of the TextOutline is the the transformations applied to a shape are also stored. This allows the Atom Symbol instances to be cached for improved efficiency if required (not currently done).
Hydrogens are positioned Above, Right, Below, or to the Left of the element label. These positions are represented and determined by the HydrogenPosition enumeration.
The hydrogen position is determined purely on the bond positions connected to each atom. When there are no bonds connected to the atom, the hydrogen label is placed depending on the element. For example it is on the left for 'H2O' and 'H2S' but on the right for 'CH4' and 'NH3'.
When a single bond is connected to the atom, the symbol is placed on the left or right.
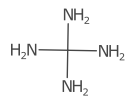
When two bonds are connected, their direction is averaged and the cardinal direction calculated. The label is placed on the opposite side, favouring the left and right positions.
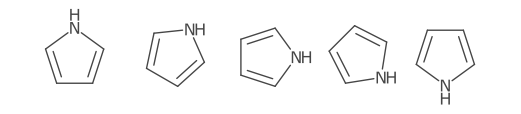
Using the cardinal direction for more than two bonds can be unsatisfactory, especially when the bond is symmetric.
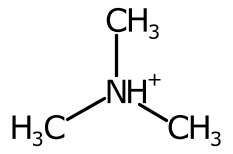
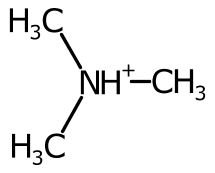
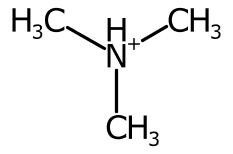
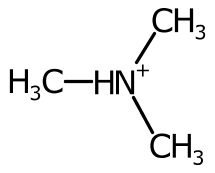
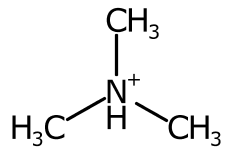
To improve the positioning, when more than two bonds are connected the sweep of the bonds around each potential position is inspected and ranked.
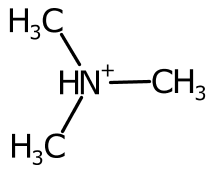
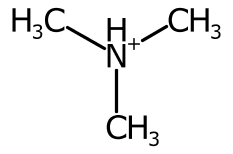
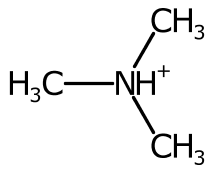
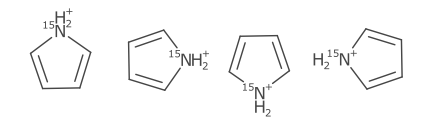
The AtomSymbol class pairs an element label with several adjuncts. The current adjuncts are hydrogen count, ionic charge, unpaired electrons, and atomic mass. The StandardAtomGenerator builds atom symbols by positioning the TextOutline of each adjunct. The relative positioning is based on the hydrogen position.
The ionic charge and unpaired electron count is placed as superscript to the top right, the atomic mass is placed to the top left (Brecher 2008). An extreme example of the adjunct positioning is shown below.
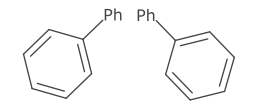
Atom symbols are aligned such that the centre of the element label lies on the point of the atom. An exception to this is when the element has multiple characters and there is exactly one bond adjacent to the atom. In this case, the atom symbol is aligned to either the first or last character.
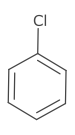
This alignment is also used for labelled pseudo atoms and aliases.
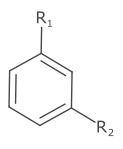
Labels that look like R group positions (R<NUMBER>+) have special treatment and position the number as subscript. Otherwise the label is rendered as the element label.
Additional support for superatoms and formal contraction is intended in future.
Aesthetically pleasing stroke widths are close to the size of the vertical or horizontal bars in the 'H' symbol (Brecher 2008). That is, the stroke should be a similar size to the font stroke.
To approximate this, the standard generator uses the width to the width of the '|' (pipe) character. The default value is '1' which means the stroke is the same width.
Using an artificial example with 18 pt Plain Verdana font shows this accomplishes a close approximation.

As this is highly dependant on the font used and so the ratio can be modified with the StrokeRatio parameter.
rendererModel.set(StandardGenerator.StrokeRatio.class,
0.8);The convex hull of the atom symbol is used to compute incoming bond intersection and back off (Clark 2013). An exaggerated example shows how the 24 incoming bonds follow the hull of this artifical 'oOo' symbol.

The amount a bond is backed off is determined by the SymbolMarginRatio. The margin is relative to the stroke width, the default margin is 2. To increase the margin:
rendererModel.set(StandardGenerator.SymbolMarginRatio.class,
4d);Single bonds without any recognized stereochemistry type are drawn as a single line between two atoms. If symbols at either end the line is offset using the convex hull intersection.
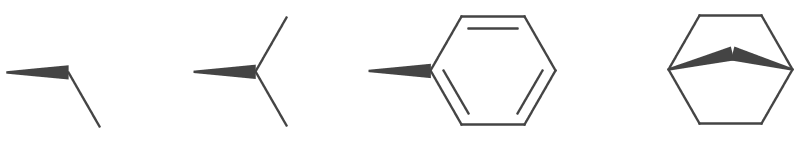
Bold wedges are drawn for IBond.Stereo.UP and IBond.Stereo.UP_INVERTED stereo types. They are depicted as a solid trapezoid, one end is the width of the stroke, the other is the stoke multiplied by the WedgeRatio. The default value is 6. The trapezoid is truncated if atom symbols are present at either or both ends.
The following hypothetical example demonstrates this:
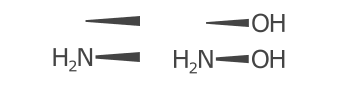
Setting a smaller wedge ratio produces less pronounced wedges.
rendererModel.set(StandardGenerator.WedgeRatio.class,
4.5d);To improve aesthetics some wedges are modified to be flush with adjacent bonds.
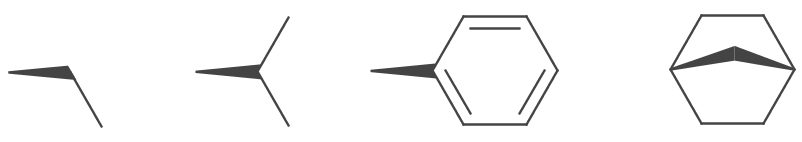
This can be turned off by disabling the FancyBoldWedges parameter.
rendererModel.set(StandardGenerator.FancyBoldWedges.class,
false);
Hashed wedges are drawn for IBond.Stereo.Down and IBond.Stereo.DOWN_INVERTED stereo types. They are depicted as a trapezoid shape overall but drawn with intermittent lines. Similar to the bold wedges, one end is the width of the stroke, the other is the stoke multiplied by the WedgeRatio. The trapezoid is truncated if atom symbols are present at either or both ends.
The following hypothetical example demonstrates this:
The spacing between the hashes is defined by the HashSpacing and BondLength parameters. The default for each of these is 5 and 40 respectively. This means that by default, ~8 hashed sections are drawn when no symbols are defined.
To render hashes with more sections the spacing can be decreased:
rendererModel.set(StandardGenerator.HashSpacing.class,
4.5);Hashes are defined by spacing rather than count to allow consistent rendering of abnormal length bonds:

For a real example, please see CHEBI:2955
To improve aesthetics the angle of hashes in some bonds is modified to be flush with adjacent bonds.
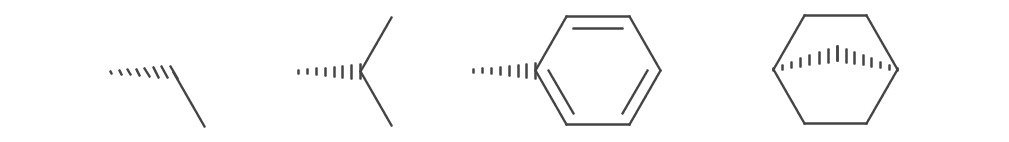
This can be turned off by disabling the FancyHashedWedges parameter.
rendererModel.set(StandardGenerator.FancyHashedWedges.class,
false);
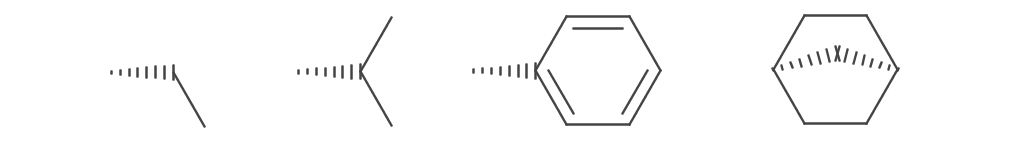
Wavy bonds are drawn for IBond.Stereo.UP_OR_DOWN and IBond.Stereo.UP_OR_DOWN_INVERTED stereo types. The bond depiction is undirected (i.e. not wedged). The wavy bond is rendered at quarter circle increments and accounts for adjacent atom symbols.
The following hypothetical example demonstrates this (note bad interpolation):
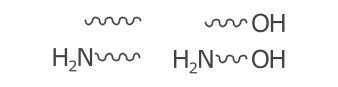
Similar to hashed wedges, the spacing between the waves is defined by the WaveSpacing and BondLength parameters. The default for each of these is 5 and 40 respectively. This means that by default, ~8 wave sections are drawn when no symbols are defined. A wave section is a half circle.
To render wavy bonds with more sections the spacing can be decreased:
rendererModel.set(StandardGenerator.WaveSpacing.class,
4.5);As with hashes, waves are defined by spacing rather than count to allow consistent rendering of abnormal length bonds:

There are currently three styles of double bonds depicted. The spacing between the lines is defined by the BondSeparation and BondLength parameters. The BondSeparation defines the percentage of bond length with
which double bonds should be separated. The default values is 18%.
To render wider bonds, the value is increased.
rendererModel.set(StandardGenerator.BondSeparation.class,
0.25);Offset double bonds are drawn with one line to the side. These bonds are depicted for asymmetric layouts and rings. The offset line is shortened to align with adjacent bonds (Brecher 2008).
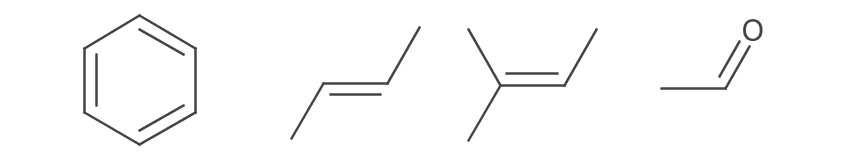
When a bond is between two rings, the offset is determined by size, number of double bonds, and finally element frequency. Rings size preference is 6 > 5 > 7 > 4 > 3 > n. If two rings are the same size the one with more double bonds is chosen. If there is still a tie, the ring with more C > N > O > S > P is chosen.
The placement is demonstrated by the following depictions:
Offset bonds are also drawn in macro cycles.
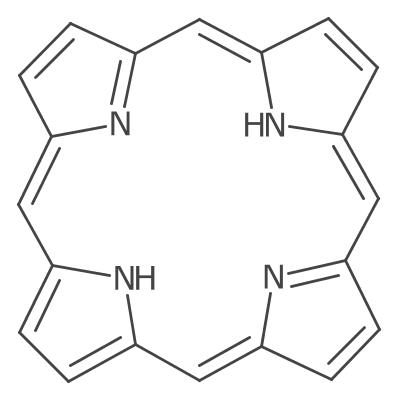
Centered double bonds draw each line equidistant from the central bond line. These are drawn when the layout is symmetric or the atom symbols at both ends are displayed. When the centered bond is next to two (or more) single bonds (e.g. ketone), the lines are lengthen to intersect with these bonds.
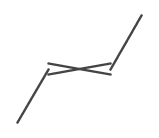
The crossed bond is depicted when the stereo type of the bond is IBond.Stereo.E_OR_Z.
Triple bonds are depicted by combining the plain single bond and a centered double bond.
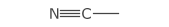
Emphasizing the atoms and bonds of a structure has be made as simple as possible. To highlight part of a substructure simply set the desired highlight color to the HIGHLIGHT_COLOR property.
IAtomContainer container = ...;
IBond bond = container.getBond(0);
IAtom atom = container.getAtom(0);
bond.setProperty(StandardGenerator.HIGHLIGHT_COLOR, Color.RED);
atom.setProperty(StandardGenerator.HIGHLIGHT_COLOR, Color.RED);Alternatively, a part of a structure can be selected/highlighted as an IChemObjectSelection.
IChemObjectSelection selection = ...; // e.g. from JChemPaint
rendererModel.setSelection(selection);
// the color is defined by the RendererModel parameter
rendererModel.set(RendererModel.SelectionColor.class, Color.BLUE); After indicating the highlight. The atoms and bonds are then highlighted in accordance with the desired highlight style.
The default highlight style is to color the atom symbols and bonds.
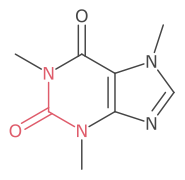
This can also be set manually as follows:
rendererModel.set(StandardGenerator.Highlighting.class,
StandardGenerator.HighlightStyle.Colored);Another available style is to add an outer glow.
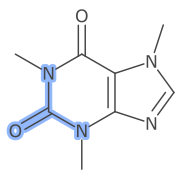
This can be enabled as follows:
rendererModel.set(StandardGenerator.Highlighting.class,
StandardGenerator.HighlightStyle.OuterGlow);The width of the glow (relative to the stroke width) can be set as follows:
rendererModel.set(StandardGenerator.OuterGlowWidth.class,
1d);As discussed in the atom generation, atom symbols are displayed depending on the Visibility parameter. The default option for highlighting is to override the display of the symbol if the atom isn't next to any highlighted bonds.
An alternative is to display the atom symbol for any highlighted atom.

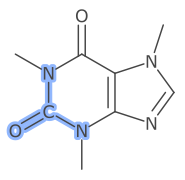
This is set by augmenting the default SymbolVisibility with a SelectionVisibility. For the example above the following parameter value is set.
// import static org.openscience.cdk.renderer.SelectionVisibility.*
// import static org.openscience.cdk.renderer.SymbolVisibility.*
rendererModel.set(StandardGenerator.Visibility.class,
all(iupacRecommendationsWithoutTerminalCarbon()));Multiple highlight colors can be used when highlights are triggered by atom properties. To demonstrate this we can bind atom mapping / atom class information to a specific highlight color. The following snippet highlights atoms based on this value.
Color[] colors = new Color[]{new Color(0xF07E82), // red (pastal)
new Color(0x98F08E), // green (pastal)
new Color(0xF0EC75)}; // yellow (pastal)
for (IAtom atom : container.atoms()) {
Integer atomMapping = atom.getProperty(CDKConstants.ATOM_ATOM_MAPPING);
// range check
if (atomMapping == null && atomMapping < 0 && atomMapping > colors.length)
continue;
// set color, atom mapping starts at '1..n'
atom.setProperty(StandardGenerator.HIGHLIGHT_COLOR,
colors[atomMapping - 1]);
}Generating a depiction for the following SMILES input is shown below.
Input: C[CH:2](C)[CH2:3][CH2:2]C(C)CCC1=C(C[CH:1]=[CH2:1])C=C[CH:2]=C1

The standard generator can include atom and bond annotation labels. The two primary use cases are atom numbering and Cahn-Ingold-Prelog descriptors. However the mechanism is generic and allows any text to be displayed.
To annotate an atom or bond simply set the StandardGenerator.ANNOTATION_LABEL property to the string label.
For example, to label to the atoms with the order they are stored:
for (IAtom atom : container.atoms()) {
// important: label must be a 'String' instance, we increment
// the number as IAtomContainer actually returns the index
String label = Integer.toString(1 + container.getAtomNumber(atom));
atom.setProperty(StandardGenerator.ANNOTATION_LABEL, label);
}The default color of the labels is Color.Red, this can be changed with the AnnotationColor parameter. Annotating in a different color is recommend to distinguish from atom symbols.
rendererModel.set(StandardGenerator.AnnotationColor.class,
new Color(0x455FFF));

The font size used is scaled relative to the atom symbol font and the distance the label is placed is relative to the bond length.
rendererModel.set(StandardGenerator.AnnotationFontScale.class,
0.4); // smaller, 40% of atom symbol font
rendererModel.set(StandardGenerator.AnnotationDistance.class,
0.15); // closer, 15% of bond lengthThe atom number need not be the order the atoms are stored. For example if a method is available to determine IUPAC numbers the following is possible.

Atom mapping and atom classes can also be used by setting the annotation label to the ATOM_ATOM_MAPPING property.
for (IAtom atom : container.atoms()) {
atom.setProperty(StandardGenerator.ANNOTATION_LABEL,
Integer.toString(CDKConstants.ATOM_ATOM_MAPPING));
}Cahn-Ingold-Prelog (CIP) stereochemistry descriptors can be labelled in a similar manner to the atom numbering. One difference is the labels are normally italic. To render labels in italics, the StandardGenerator.ITALIC_DISPLAY_PREFIX can be used. This property is added as prefix to the annotation label.
For example - to add CIP R/S and E/Z labels to stereogenic atoms and bonds.
CIPTool.label(container);
for (IAtom atom : container.atoms())
atom.setProperty(StandardGenerator.ANNOTATION_LABEL,
getCipLabel(atom));
for (IBond bond : container.bonds())
bond.setProperty(StandardGenerator.ANNOTATION_LABEL,
getCipLabel(bond));
// helper method
static String getCipLabel(IChemObject chemObj) {
String label = chemObj.getProperty(CDKConstants.CIP_DESCRIPTOR);
if (label == null) return null;
return StandardGenerator.ITALIC_DISPLAY_PREFIX + label;
}
