This repo was made by Minh H. Vu based on an amazing work for MUTAN. We developed this code in the frame of a research paper called A Question-Centric Model for Visual Question Answering in Medical Imaging which is the current state-of-the-art on the medical images.
The goal of this repo is two folds:
- to make it easier to reproduce our results,
- to provide an efficient and modular code base to the community for further research on other medical VQA datasets.
If you have any questions about our code or model, don't hesitate to contact us or to submit any issues. Pull request are welcome!
- 29/02/2020: paper accepted at IEEE Transactions on Medical Imaging
The task is about training models in a end-to-end fashion on a multimodal dataset made of triplets:
-
an image with no other information than the raw pixels,
-
a question about visual content(s) on the associated image,
-
a short answer to the question (one or a few words).
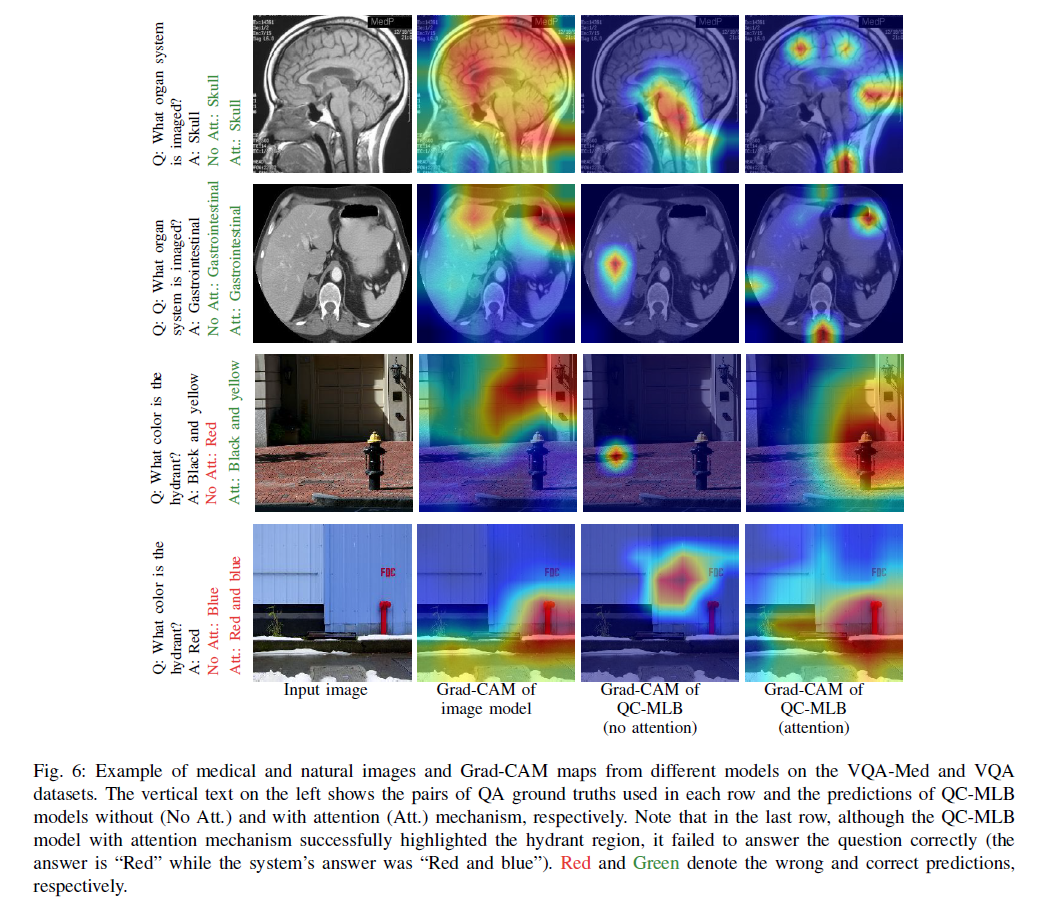
As you can see in the illustration bellow, two different triplets (but same image) of the VQA dataset are represented. The models need to learn rich multimodal representations to be able to give the right answers.
The VQA task is still on active research. However, when it will be solved, it could be very useful to improve human-to-machine interfaces (especially for the blinds).
The VQA community developped an approach based on four learnable components:
-
a question model which can be a LSTM, GRU, or pretrained Skipthoughts,
-
an image model which can be a pretrained VGG16 or ResNet-152,
-
a fusion scheme which can be an element-wise sum, concatenation, MCB, MLB, or Mutan,
-
optionally, an attention scheme which may have several "glimpses".
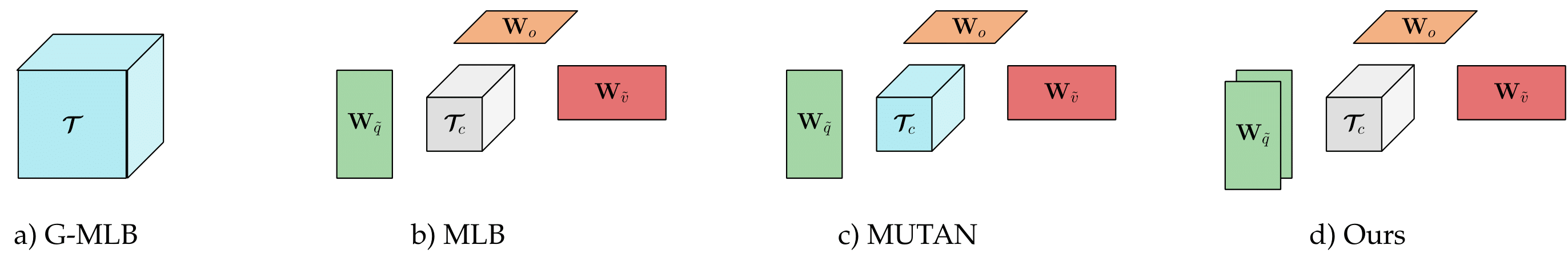
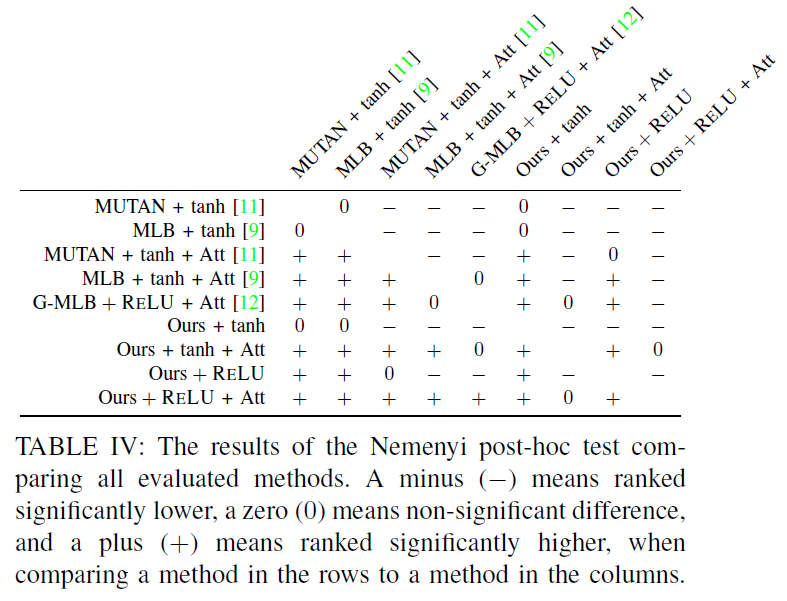
One of our claim is that the multimodal fusion between the image and the question representations is a critical component. Thus, our proposed model uses a Tucker Decomposition of the correlation Tensor to model richer multimodal interactions in order to provide proper answers. Our best model is based on :
- a pretrained Skipthoughts for the question model,
- features from a pretrained Resnet-152 (with images of size 3x448x448) for the image model,
- our proposed Mutan (based on a Tucker Decomposition) for the fusion scheme,
- an attention scheme with two "glimpses".
First install python 3 (we don't provide support for python 2). We advise you to install python 3 and pytorch with Anaconda:
conda create --name vqa python=3
source activate vqa
conda install pytorch torchvision cuda80 -c soumith
Then clone the repo (with the --recursive flag for submodules) and install the complementary requirements:
cd $HOME
git clone --recursive https://github.com/Cadene/vqa.pytorch.git
cd vqa.pytorch
pip install -r requirements.txt
Our code has two external dependencies:
-
VQA is used to evaluate results files on the valset with the OpendEnded accuracy,
-
skip-thoughts.torch is used to import pretrained GRUs and embeddings,
-
pretrained-models.pytorch is used to load pretrained convnets.
Data will be automaticaly downloaded and preprocessed when needed. Links to data are stored in vqa/datasets/vqa.py, vqa/datasets/coco.py and vqa/datasets/vgenome.py.
.
├── options # default options dir containing yaml files
├── logs # experiments dir containing directories of logs (one by experiment)
├── data # datasets directories
| ├── coco # images and features
| ├── vqa # raw, interim and processed data
| ├── vgenome # raw, interim, processed data + images and features
| └── ...
├── vqa # vqa package dir
| ├── datasets # datasets classes & functions dir (vqa, coco, vgenome, images, features, etc.)
| ├── external # submodules dir (VQA, skip-thoughts.torch, pretrained-models.pytorch)
| ├── lib # misc classes & func dir (engine, logger, dataloader, etc.)
| └── models # models classes & func dir (att, fusion, notatt, seq2vec, convnets)
|
├── train.py # train & eval models
├── eval_res.py # eval results files with OpenEnded metric
├── extract.py # extract features from coco with CNNs
└── visu.py # visualize logs and monitor training
There are three kind of options:
-
options from the yaml options files stored in the
optionsdirectory which are used as default (path to directory, logs, model, features, etc.) -
options from the ArgumentParser in the
train.pyfile which are set to None and can overwrite default options (learning rate, batch size, etc.) -
options from the ArgumentParser in the
train.pyfile which are set to default values (print frequency, number of threads, resume model, evaluate model, etc.)
You can easly add new options in your custom yaml file if needed. Also, if you want to grid search a parameter, you can add an ArgumentParser option and modify the dictionnary in train.py:L80.
We currently provide four datasets:
-
COCOImages currently used to extract features, it comes with three datasets: trainset, valset and testset
-
VisualGenomeImages currently used to extract features, it comes with one split: trainset
-
VQA 1.0 comes with four datasets: trainset, valset, testset (including test-std and test-dev) and "trainvalset" (concatenation of trainset and valset)
-
VQA 2.0 same but twice bigger (however same images than VQA 1.0)
Please cite this paper if you use our work:
@ARTICLE{9024133,
author={M. H. {Vu} and T. {Löfstedt} and T. {Nyholm} and R. {Sznitman}},
journal={IEEE Transactions on Medical Imaging},
title={{A Question-Centric Model for Visual Question Answering in Medical Imaging}},
year={2020},
volume={39},
number={9},
pages={2856-2868},
doi={10.1109/TMI.2020.2978284}}
This research was conducted using the resources of the High Performance Computing Center North (HPC2N) at Umeå University, Umeå, Sweden. We are grateful for the financial support obtained from the Cancer Research Fund in Northern Sweden, Karin and Krister Olsson, Umeå University, The Västerbotten regional county, and Vinnova, the Swedish innovation agency.