This repository includes code for NER and RE methods on EHR records. These methods were performed on n2c2 2018 challenge dataset which was augmented to include a sample of ADE corpus dataset. This project serves as a capstone project for my Masters in Data Science degree at Northeastern University. A demo of this project can be accessed at ehr-info.ml. The website might not work if the GCP instance is turned off (it costs a lot of money, especially for a student).
- How to Run
- Introduction
- Named Entity Recognition
- Relation Extraction
- End-to-End Results
- Front-end and API deployment
- References
Edit the Makefile for any parameter changes that you want. All parameters are defined at the top of the file. Check expected parameter values in the next section.
- Generate data:
make generate-data - Train BioBERT for NER:
make train-biobert-ner - Train BiLSTM + CRF for NER:
make train-bilstm - Train BioBERT for RE:
make train-biobert-re - Run API in development mode with debugging:
make start-api-local - Run API in production mode:
make start-api-gcp - Run the front-end: Edit the IP address for AJAX call in
front-end/ehr.htmland open the HTML file in a browser.
-
To generate the preprocessed data required for model input
python generate_data.py \ --task ner \ --input_dir data/ \ --ade_dir ade_corpus/ \ --target_dir dataset/ \ --max_seq_len 512 \ --dev_split 0.1 \ --tokenizer biobert-base \ --ext txt \ --sep " " \- The
taskparameter can be eithernerorrefor Named Entity Recognition and Relation Extraction tasks respectively. - The input directory should have two folders named
trainandtestin them. Each folder should havetxtandannfiles from the original dataset. ade_diris an optional parameter. It should contain json files from the ADE Corpus dataset.- The
max_seq_lenshould not exceed512for BioBERT models. - For BioBERT models, use
biobert-baseas thetokenizervalue and for BiLSTM + CRF model, usescispacy_plus. - Use
txtfor theext(extension) parameter and" "as thesep(seperator) parameter for NER, andtsvextension andtabas the seperator for RE.
- The
-
Instructions for running individual models can be found in their respective directories.
-
To run the API in development mode with debugging on, run the following command:
uvicorn fast_api:app --reload -
To run the API in production mode with gunicorn, run the following command:
gunicorn -b 0.0.0.0:8000 -w 4 -k uvicorn.workers.UvicornWorker fast_api:app --timeout 120 -
To run the front-end, edit the IP address for AJAX call in
front-end/ehr.htmland open the HTML file in a browser.
An Electronic Health Record (EHR) [1] is an electronic version of a patient's medical history that includes extremely important information including, but not limited to, problems, medication, progress notes, immunizations and laboratory reports. EHRs are huge free-text data files that are documented by healthcare professionals, like clinical notes, discharge summaries or lab reports. Finding information from this data is time consuming, since the data is unstructured and there may be multiple such records for a single patient. Natural Language Processing (NLP) techniques could be used to make this data structured, and quickly find information whenever needed, thereby saving healthcare professionals' time from these mundane tasks.
In this project, we aim to build a tool that would automatically structure this data into a format that would enable doctors and patients to quickly find information that they need. Specifically, we aim to build a Named Entity Recognition (NER) model that would recognize entities such as drug, strength, duration, frequency, adverse drug event (ADE) [2], reason for taking the drug, route and form. Further, the model would also recognize the relationship between drug and every other named entity as well. This would allow healthcare professionals to not only look at individual entities, but also all the relationships between them. This would also allow the doctors to easily find out the relationships between a drug and ADEs so that such drugs can be monitored carefully. \par
The final goal of this project is to build an API where healthcare professionals and patients could send EHR data and the API would return character ranges for each annotation so they can be highlighted in the original data, a structured json-format data that includes separately labelled data for medication history and discharge medications. The highlighted annotations could be useful when a healthcare professional wants to see important information along with other details in the EHR. The structured information can be used to store the data for quick reference in the future. Because the EHR contains medication history as well as discharge medications, labelling them as such could help in merging new information, as the medication history would remain the same.
To identify named entities from the text, three different models were built. A rule-based model was built as a baseline along with two machine learning models.
EHR documents are usually lengthy, and it is not desirable to have such big input sizes for machine learning models, especially for models like BERT that have an input size restriction of 512 tokens. So, a function was implemented that would split the EHR records based on a maximum sequence length parameter. The function tries to include maximum number of tokens, maintaining as much context as possible for every token. The splitting points are decided based on the following criteria:
- Includes as many paragraphs as possible within the maximum token limit, and splits at the end of the last paragraph found.
- If the function cannot find a single complete paragraph, it splits on the last line (within the token limit) which marks the end of a sentence.
- Otherwise, the function includes as many tokens as specified by the token limit, and then splits on the next index.
The data is tokenized using a modified ScispaCy tokenizer for BiLSTM + CRF model which just removes the tokens with whitespace characters after ScispaCy tokenizes them. For BioBERT model, the BioBERT base tokenizer was used to tokenize the data. Each sequence of labels or tokens in the data was represented using the IOB2 (Inside, Outside, Beginning) tagging scheme for BioBERT and BiLSTM models.
To establish a baseline, a traditional dictionary and regular-expression based NER model was used. A regular expression was written to find the dosage entity, which would find any number followed by "mg" or "mcg". For all other entities, the data was split into 80% train data and 20% test data. The train data was used to create a dictionary of each entities, so if the same entities appear in the test data, it would classify it as the corresponding entity.
Just a BiLSTM network is enough to classify each token into various entities along with it's class (i.e. B: beginning or I: inside) or if it is not a part of any of the entities we are looking for (O: outside) but we witnessed some common errors of misclassification. Because the outputs of BiLSTM of each word are the label scores, we can select the label which has the highest score for each word. By this scheme, we may end up with invalid outputs, for eg: I-Drug followed by I-ADE or B-Drug followed by I-ADE. Hence we use the CRF (Conditional Random Field) algorithm to calculate the loss of our BiLSTM network as it could add some constraints to the final predicted labels to ensure they are valid. These constraints can be learned by CRF automatically from the training dataset during the training process. CRFs considers the context as well rather than predicting label for a single token without considering neighboring samples [3].
The model was built using the architecture described in Guillaume Genthial's Blog [4] and it's PyTorch implementation [5]. To train the model, the EHR dataset was tokenized using a modified version of the ScispaCy [6] tokenizer. The original tokenizer keeps all whitespace characters in separate tokens, but it was modified so that all of the white space tokens are removed. Every other tokens would remain the same. The input sequence length was set to 512 and the EHR records were split by using the steps discussed in the preprocessing section. The model was trained for 15 epochs using GPU resources from Google Cloud compute engine.
The output of each token from the BERT model is passed through a fully connected neural network with a softmax layer at the end that classify that token to an entity. The entities here would be in IOB format, for example B-DRUG and I-DRUG would be treated as separate entities. This entire model is called BERT for token classification, and it's architecture is available in python's transformers library [7].
BioBERT is a pre-trained BERT model, that is trained on medical corpra of more than 18 billion words. Since it has a medical vocabulary and is trained on biomedical data, we chose this model to fine tune on our dataset. Code for fine tuning from the official BioBERT for PyTorch GitHub repository [8] was used with modifications in input format. The input sequence length was set to 128, and the model was fine tuned for 5 epochs using GPU resources from Google Cloud compute engine.
The Rule-Based model did not perform very well, but that was expected as it does not take context into account and has a very high false positive rate. For BiLSTM + CRF and BioBERT, a sample of an external dataset, the ADE-corpus dataset\cite{ade-corpus} was integrated to our data which improved the performance to a great extent. The F1 score for ADE entity improved from 0.3403 to 0.8673 for the BioBERT model after adding a sample of the ADE Corpus.
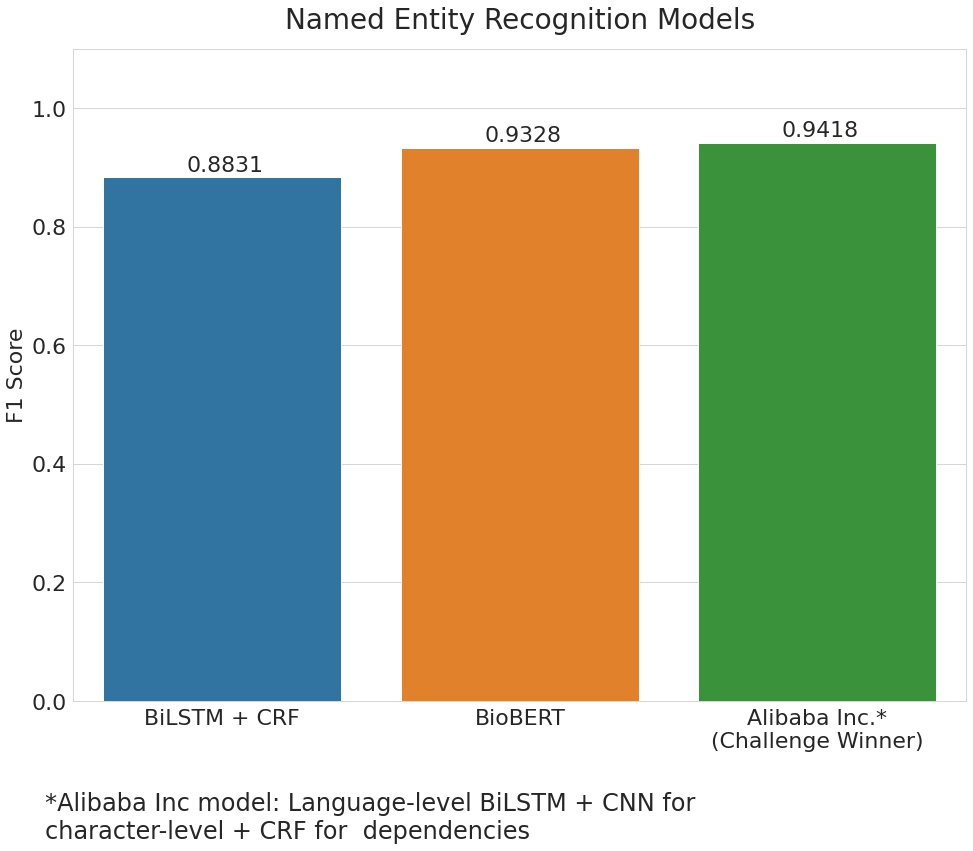
Also, the BioBERT and BiLSTM + CRF models produce F1 scores similar to that of the model that won the n2c2 challenge using the same data-set. The winning model was submitted by Alibaba Inc. and used an architecture of BiLSTM + CNN for character-level + CRF for dependencies.
| Model | Micro F1 |
|---|---|
| Rule Based | 0.2200 |
| BiLSTM + CRF | 0.8831 |
| BioBERT | 0.9328 |
| Alibaba Inc. (Challenge winner) | 0.9418 |
For the relation extraction task, a BERT for sequence classification model was used.
Similar to the NER model, in order to be able to train and test the RE model, the data had to be transformed in a particular format. After splitting the train data into train and dev, each record was further split into paragraphs using the same method that was used for NER. The next step was to map each drug entity with all the other possible entities within that paragraph. This would form a list of all possible relations in that paragraph. Once the list was obtained, each entity text was replaced with @entity-type$. For example, the drug 'Lisinopril' would be replaced by @Drug\$ and '20mg' with @Strength\$. This was done for each relation, which means each data point would have only one relation i.e. one pair of entities. Finally, a label tag that indicates whether the entities in that text are related was added - 1 representing a relationship and 0 otherwise.
When taken a close look at the transformed data, Relation Extraction (RE) is nothing but a binary classification problem. It was decided to use the BioBERT model again as the training process was much faster when compared to the LSTM models. Also, the biomedical domain knowledge would be an advantage over other methods.
Unlike NER where it was used for token classification, BioBERT uses the concept of sequence classification in order to predict the relations. In sequence classification, a sentence-level representation of the input sentence, called the CLS (stands for classification) token is obtained. This CLS token which basically contains both word-level and contextual information of the whole sentence, is fed into a fully connected neural network which implements the binary classification task. This model was trained on the same specifications as the NER BioBERT model.
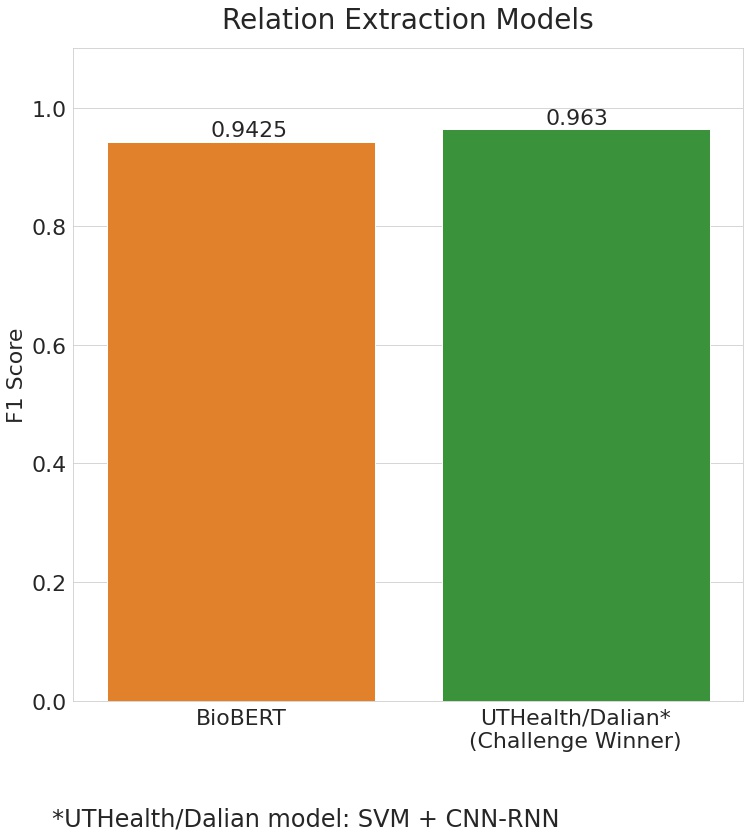
Even for the purpose of RE, the BioBERT model seemed to have performed extremely well. With an overall F1 score of 0.942, the model was short of just 0.021 when compared to the challenge winners' score which was of 0.963.
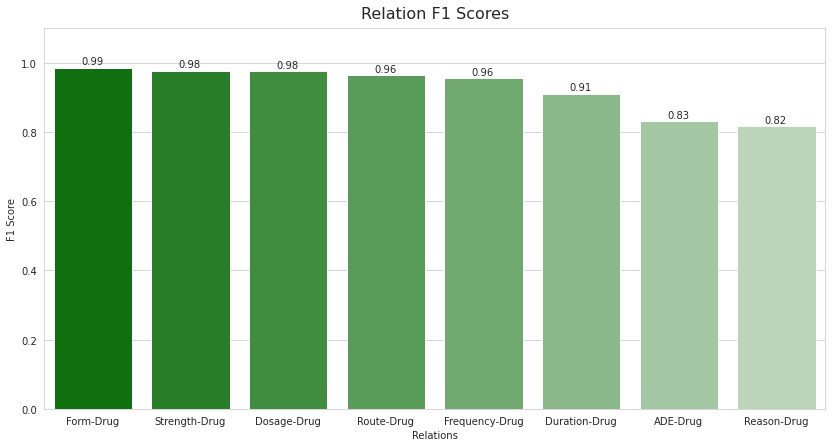
In addition to this, the model managed to achieve high F1 scores for each type of relation as well. The highest being that of Form-Drug with a score of 0.99, followed by Strength-Drug and Dosage-Drug, each with a score of 0.98. It is interesting to see that the scores for both the Reason-Drug and ADE-Drug relations are similar with 0.82 and 0.83, respectively. This also suggests that adding the external ADE corpus seemed to have improved the F1 scores for the ADE-Drug relation (though conclusive proof was not obtained as the model was not trained without the ADE corpus). Also, based on these results, it can be said that the model is doing a decent job of differentiating between the above two relations which is a crucial part of this project.
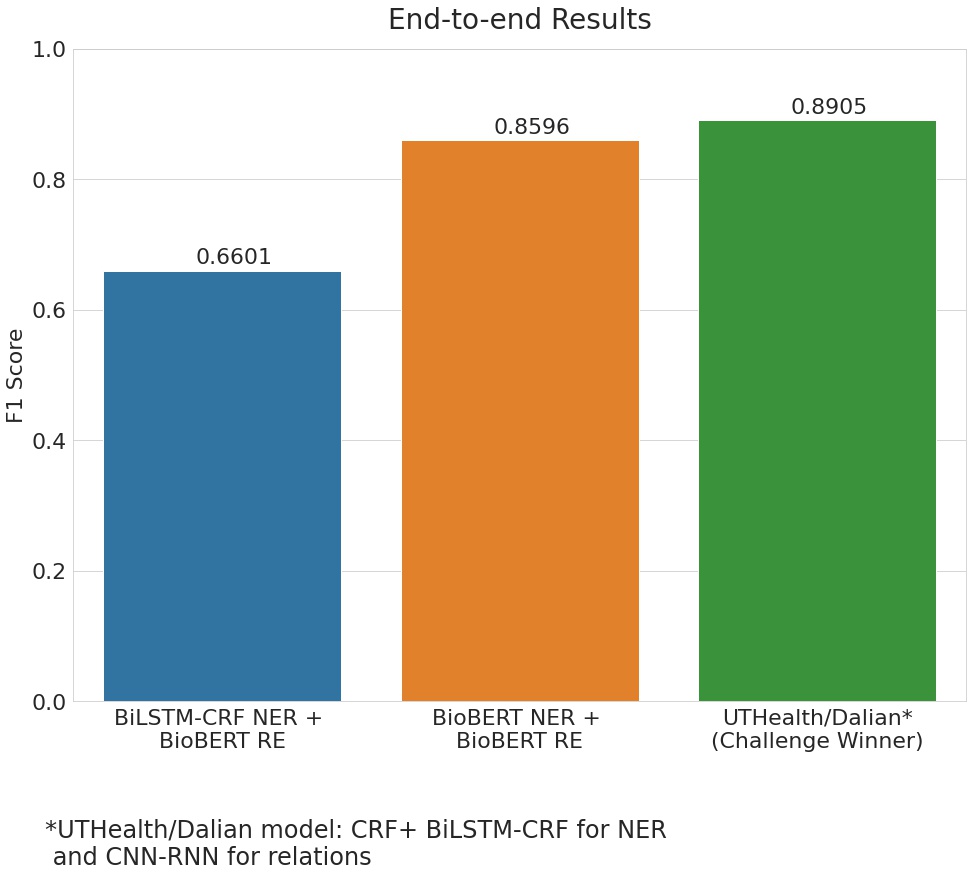
The above Relation Extraction results were obtained when using the actual entities provided in the data. But in reality, this will not be the case since the relationships would be obtained using the entities predicted by the NER model. This is what the end-to-end pipeline represents and the corresponding scores can be seen in the figure below. By using the BioBERT model for both Named Entity Recognition and Relation Extraction, we get an F1 score of 0.86 which is a significant drop from the earlier score of 0.94. The obvious reason for this reduction is the cascading effect of the entities that were incorrectly predicted by the NER model. However, it should be noted that the challenge winners' experienced a drop in the F1 scores as well. Moreover, the drop is even more significant when using the BiLSTM + CRF model for predicting the entities.
An API was built for better accesibility and a front-end website was built to showcase the work and to make it easier for users to visualize the results. They were deployed on Google Cloud Platform (GCP).
An end-to-end pipeline was created to transform raw EHR documents into a structured, and more intuitive form. First, the raw EHR document is preprocessed for a Named Entity Recognition model. This preprocessed data is then sent passed through either BiLSTM + CRF model or BioBERT model for NER based on user's choice, to get predictions of entities present in the EHR document. Using these predicted entities, the raw EHR is again preprocessed for the Relation Extraction model. After getting the predictions of relationships among the entities, a table and a knowledge graph is generated which maps each drug to all of its related entities. The relation table is created using the python pandas package and the knowledge graph is created using python's networkx package.
This end-to-end pipeline was converted into an API using a python web-framework named FastAPI [9]. This package allows building a production-ready API and is compatible with HTTP web servers like Gunicorn [10].
To visualize the results of the end-to-end pipeline, a static front-end website was built using HTML, Bootstrap and jQuery. The Bootstrap framework ensures that the website layout changes automatically if the website is accessed on a mobile device. The website provides an option to choose the NER model and an option to either upload or type/paste an EHR document. It also provides the user with an option to load a sample EHR document for the user to test the results. Once the user requests for the results, an AJAX [11] call is made to the API which sends the EHR text and the NER model choice to the API. The API then runs the entire pipeline and transfers the results to the website.
To visualize the results and make it more user friendly, the website highlights each entity with a different color and hovering over the highlighted text gives a tooltip of the entity type. To visualize the relations of an entity, hovering over an entity creates a red colored border around all other entities that are related to it. The website also has an option to visualize all the relations in a tabular format and in a knowledge graph.
The API was first deployed on Google Cloud Platform's App Engine service, which provides Platform as a Service (PaaS) where the application can be managed without the complexity of building infrastructure like networks, servers, operating system and such. We just have to submit the deployment code and everything is managed automatically. However, App Engine does not currently support GPUs which caused the run-times of the models to be very high.
Due to the lack of GPU support in App Engine, the API and the front-end website was then deployed on GCP Compute Engine, which provides Infrastructure as a Service (IaaS). On Compute Engine, everything from installing an operating system, building a web server, managing networks, obtaining certificates and domain names, and managing load balancing was done by us. The final website can be accessed on ehr-info.ml
[1] Electronic Health Records: https://www.cms.gov/Medicare/E-Health/EHealthRecords
[2] Adverse Drug Events: https://www.cdc.gov/medicationsafety/adult_adversedrugevents.html
[3] Conditional Random Fields: https://medium.com/ml2vec/overview-of-conditional-random-fields-68a2a20fa541
[4] Guillaume Genthial's Blog: https://guillaumegenthial.github.io/
[5] PyTorch-ELMo-BiLSTM-CRF implementation: https://github.com/yongyuwen/PyTorch-Elmo-BiLSTMCRF
[6] ScispaCy: https://allenai.github.io/scispacy/
[7] HuggingFace Transformers: https://huggingface.co/transformers/
[8] DMIS Lab - BioBERT PyTorch: https://github.com/dmis-lab/biobert-pytorch/tree/master/named-entity-recognition
[9] FastAPI: https://fastapi.tiangolo.com/
[10] Gunicorn: https://gunicorn.org/
[11] jQuery AJAX Method: https://api.jquery.com/Jquery.ajax/