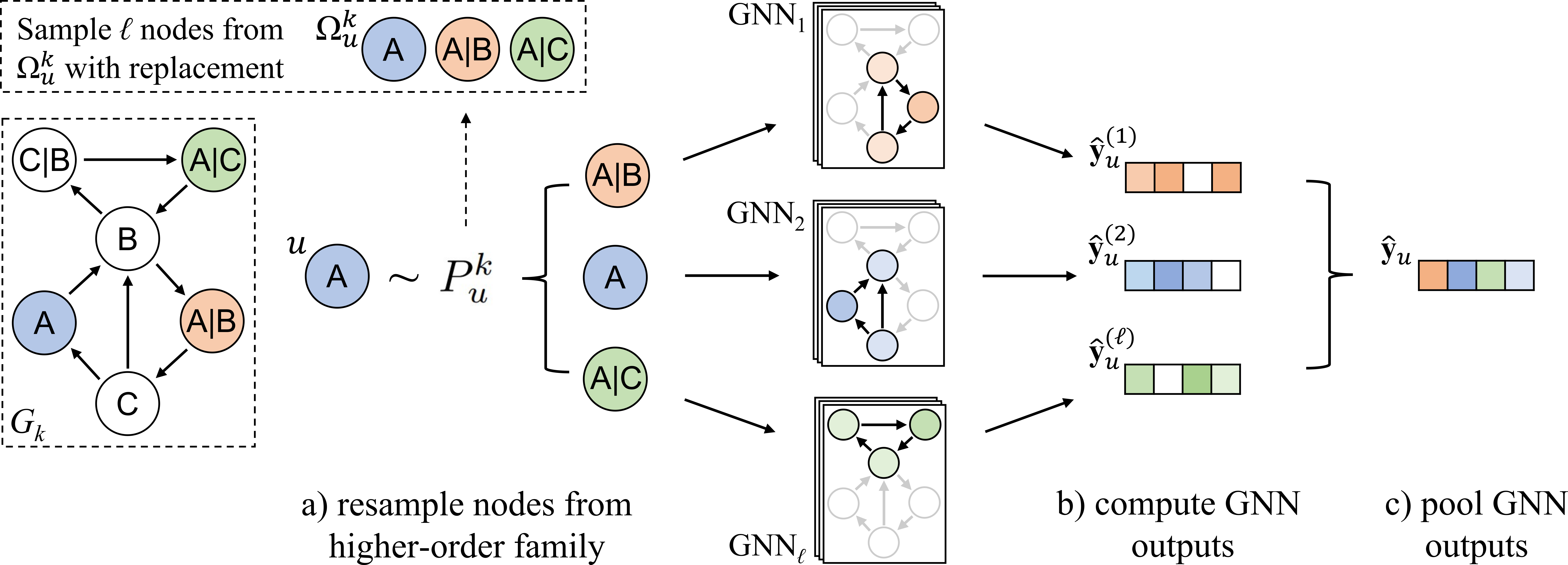
This repository contains initial code for the paper "Deep Ensembles for Graphs with Higher-order Dependencies," published in ICLR 2023. The proposed model, DGE (Deep Graph Ensembles), utilizes an ensemble of graph neural networks (GNNs) to exploit neighborhood variance in higher-order networks. In graphs with higher-order dependencies, DGE consistently outperforms all state-of-the-art baselines.
0.1 (CURRENT) Supports node classification experiments for DGE, GraphSAGE, and GIN on three of the data sets (Air, Wiki, and Ship). Please be advised that the current version of this code serves primarily to replicate the results in the paper. We plan to improve this repository to include a more modular and extensible version of the StellarGraph implementation, as well as a PyTorch/Geometric implementation.
- Python 3.7.3+
- tensorflow (tested with 2.4.1)
- StellarGraph (tested with 1.2.1)
- utils.py (misc utilities)
- sgmods.py (mods for stellargraph classes)
python dge.py {INFPREFIX}
where {INFPREFIX} is the prefix (including folder path) for the input data set. At minimum, the code expects to find an edgelist at "{INFPREFIX}{k}.txt" for the input graph of order k. Additionally, for node classification the code expects to find a file containing node labels at "{INFPREFIX}_labels.csv".
The results from each experiment be written to an output directory (./results/ by default). You can run "agg_results.py" to combine the results from multiple expeirments into a single CSV.
-task --task (int, default=0, choices={0}): 0 for node classification, 1 for link prediction (will be supported soon).
-k --k (int, default=2): max order for input graph, this is primarily used to locate the input file.
-l --learners (int, default=16): number of base GNNs for the ensemble
-p --pool (int, default=0, choices={0,1,2,3}): 0 for dge-concat (eq. 5a), 1 for dge-pool (eq. 5b)), 2 for dge-bag (eq. 5c), 3 for dge-batch*
-q --shareparams (bool, default=False): whether base learners should share params (if pool=3 this is forced to True)
-n --neighborsamples (list of int, default=[128,1]): number of neighbors to sample at each layer (e.g., the default samples 128 one-hop neighbors, and 1 two-hop neighbor for each of the sampled one-hop neighbors)
-a --aggregator (int, default=0, choices={0,1,2}): neighborhood aggregation function; 0 for mean, 1 for sum, 2 for max
-s --seed (int, default=7): random seed
-f --testfold (int, default=0, choices={0,1,2,3,4}): current testing fold for 5-fold cross validation
-d --hiddendims (list of int, default=[128,128]): hidden layer sizes per base learner; length of this list must match length of --neighborsamples
-x --dropout (float, default=0.4): GNN dropout rate
-b --batchsize (int, default=16): minibatch size
-e --epochs (int, default=25): number of training iterations for each base learner (this should vary with your choice of --pool)
-r --learningrate (float, default=0.01): learning rate
-t --tau (int, default=0): used to specify a different HON for input
-o --outdir (str, default='results/'): output directory for experimental results
You can reproduce the results of the paper with the following configurations (each needs to repeat 5-fold cross validation with -f = {0,1,2,3,4}). For example,
DGE-bag:
python dge.py dat/air
python dge.py dat/wiki -n 64 1
DGE-pool*:
python dge.py dat/air -p 1 -q
python dge.py dat/wiki -p 1 -q -n 64 1
For baselines set -k to 1 and -l to 1.
GraphSAGE:
python dge.py dat/air -k 1 -l 1 -e 50
python dge.py dat/air -k 1 -l 1 -n 64 1 -e 50
GIN:
python dge.py dat/air -a 1 -k 1 -l 1 -e 50
python dge.py dat/air -a 1 -k 1 -l 1 -n 64 1 -e 50
- One network file for each k, located at {INFPREFIX}{k}.txt (e.g. dat/toy1.txt). These must be provided in a weighted edgelist format delimited by a space. Node IDs can be strings or integers. You can use GrowHON to construct a graph with k > 1 from a set of input sequences.
- For the node classification task, a list of node labels in .csv format located at {INFPREFIX}_labels.csv. The values in first column must match the node labels in the first order graph, e.g. toy1.txt. The .csv file must contain a column called "Label" that has a non-missing value for every node in the first-order graph. There can be any number of classes and the labels can be integers or strings.
If you use this code, you can cite our paper as follows:
@inproceedings{
krieg2023deep,
title={Deep Ensembles for Graphs with Higher-order Dependencies},
author={Steven Krieg and William Burgis and Patrick Soga and Nitesh Chawla},
booktitle={The Eleventh International Conference on Learning Representations},
year={2023},
url={https://openreview.net/forum?id=hZftxQGJ4Re}
}