|
|
- What does myBrain-Seq do?
- Using the myBrain-Seq image in Linux
- Test data
- Publications
- For Developers
- Team
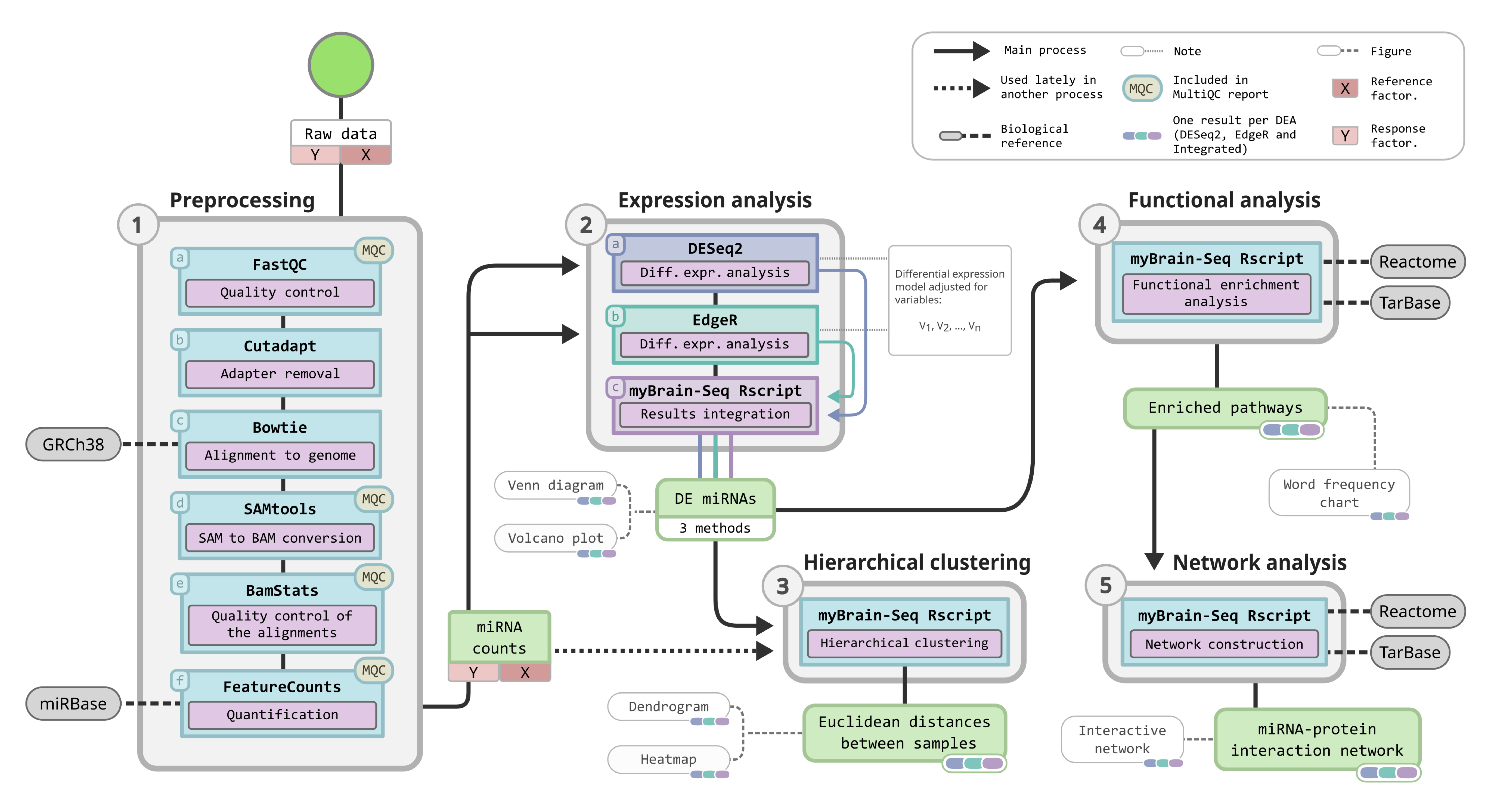
MyBrain-Seq is a Compi pipeline for performing full analyses of miRNA-Seq data, with particular interest on neuropsychiatric data.
It can automatically identify differentially expressed microRNAs (DE miRNAs) between two conditions using two differential expression analysis software, namely DESeq2 and EdgeR, and is able to offer an integrated result suitable for experimental validation. Additionally, a functional analysis module puts biological meaning behind the list of DE miRNAs and eases the process of biomarker identification.
Its features and analysis are designed and tuned to work with miRNA data. We designed myBrain-Seq with the particularities of neuropsychiatric data in mind. In this way, myBrain-Seq addresses its most common limitations while offering results that help the investigator to identify potential biomarkers and molecular mechanisms for the studied conditions. When more than two conditions are involved, myBrain-Seq facilitates performing all the pairwise comparisons of interest.
A typical analysis with myBrain-Seq comprises the following steps, which are further detailed below:
- Preprocessing
- Differential expression analysis
- Functional analysis
- Results summarization
Prepare the input FastQ files for the differential expression analysis. This process comprises:
- Quality control of the sequences using FastQC.
- Trimming of the adapter sequences using Cutadapt (optional).
- Alignment to the reference genome with Bowtie.
- Format conversion of the Bowtie output files to BAM using sam-tools.
- Quality control of the alignments with sam-tools.
- Quantification and annotation with featureCounts.
After the preprocessing was completed, myBrain-Seq performs the differential expression analysis. This process comprises:
- Differential expression analysis with DESeq2 (with/without factor correction).
- Differential expression analysis with EdgeR (with/without factor correction).
- Intersection of the DESeq2 and EdgeR results and averagement of their q-values and fold change optional).
- Creation of a venn diagram with the integrated results using VennDiagram.
- Creation of a volcano plot with the results using EnhancedVolcano.
In addition, the user can instruct myBrain-Seq to generate a genome index for the Bowtie alignment; this index will be built in parallel with the preprocessing tasks.
After the differential expression analysis, myBrain-Seq performs a functional analysis. This process comprises:
- Hierarchical clustering of the samples using the expression of the DE miRNAs.
- Functional enrichment analysis of the DE miRNAs using Diana Tarbase and Reactome databases as reference.
- Creation of a miRNA-target network, expanded using Reactome protein-protein interactions.
Finally, a single MultiQC report is generated to summarize the results of the quality, alignment, assignment and quantification of all the samples.
To perform a myBrain-Seq analysis users must first:
- Initialize a working directory with the files required myBrain-Seq.
- Add the data analysis (fastQ reads, genomes, contrast files, and so on).
- Configure the pipeline parameters.
This section provides a comprehensive guide on how to perform these steps and the tools and scripts included in the myBrain-Seq image to do it easily.
Some steps on the preparation of myBrain-Seq analysis require either to adapt and run code on a console or to use myBrain-Seq's terminal user interface (v.console). As the v.console can perform several operations, please refer to this section whenever you need to use it. To launch the v.console just run the following command on a terminal:
docker run --rm -it -v /var/run/docker.sock:/var/run/docker.sock -v /tmp:/tmp singgroup/my-brain-seq visual_console.shAn interactive menu should be displayed in your terminal.
To start a new analysis, the first thing to do is build the directory tree in your local file system. This directory tree will be referred as the working directory and its structure is recognized and used by the pipeline during the analysis.
MyBrain-seq offers two options to generate the working directory: interactively using myBrain-Seq's terminal user interface (v.console) or adapting and running a command in the console.
Run the v.console (see section "Running the v.console") and select the option "Initialize the working-directory"; then, paste the full path where the "working-directory" should be placed and confirm.
To build the working directory adapt the first line of the following code and run it:
WORKING_DIRECTORY=/path/to/the/working-directory
docker run --rm -v ${WORKING_DIRECTORY}:${WORKING_DIRECTORY} -u "$(id -u)":"$(id -g)" singgroup/my-brain-seq init_working_dir.sh ${WORKING_DIRECTORY}After completing any of the above options, the selected working-directory (mbs_project in the example below) should have the following structure:
/home/user/mbs-project
|-- README.txt
|-- input
| |-- compi.parameters
| |-- conditions_file.txt
| `-- contrast_file.txt
|-- output
`-- run.sh
Where:
- README.txt contains the next steps you need to do to run the analysis.
- compi.parameters contains the paths and parameters needed for the analysis.
- conditions_file.txt contains the names and conditions of each fastQ file.
- contrast_file.txt contains the names and labels of the conditions to compare in the differential expression analysis.
- run.sh is the script to run the analysis.
The creations of these files is detailed in the following sections as well as briefly indicated in the README.txt file. You may find it convenient to create additional directories and files within the working directory to group all the data related to a particular study.
The compi.parameters file is used by myBrain-Seq to locate the files needed for the analysis as well as to define which optional tasks will be run. Here is an example of a compi.parameters file using the working directory created in the previous example:
workingDir=/path/to/mbs-project
fastqDir=/path/to/study_1/data/
gffFile=/path/to/study_1/refs/mirbase_hsa.gff3
conditions=/path/to/mbs-project/input/conditions_file_study_1.txt
contrast=/path/to/mbs-project/input/contrast_file_study_1.txt
bwtIndex=/path/to/study_1/refs/bowtie-index_GRCh38/GCA_000001405.15_GRCh38_no_alt_analysis_set
adapter=TGGAATTCTCGGGTGCCAAGG
organism=Homo sapiens
The following are mandatory parameters:
- workingDir: the path to the myBrain-Seq working directory of the analysis (first example of this section).
- fastqDir: the path to the directory with the fastQ files to analyse.
- gffFile: the path to the the GFF3 file with the miRNA annotations. This file could be obtained from miRBase or NCBI Genomes.
- conditions: the path to conditions_file.txt.
- contrast: the path to contrast_file.txt.
- genome (optional if bwtIndex is provided): the path to the reference genome in FASTA from which the Bowtie index will be built.
- bwtIndex (optional if genome is provided): the path to a directory containing a Bowtie index, including the basename of the bowtie index files. If this parameter is omitted myBrain-Seq will build a new index using a genome in FASTA provided in the genome parameter.
- organism: the organism used in the study. This parameter is used for the functional enrichment analysis and for the network construction. Available organisms are: Mus musculus, Homo sapiens, Caenorhabditis elegans, Danio rerio, Rattus norvegicus, Gallus gallus, Drosophila melanogaster.
Whereas these are some useful optional parameters:
- adapter: the sequence of the adapter to remove. If this parameter is omitted myBrain-Seq will skip the adapter removal step.
- gffFeature: the name of the feature of the GFF3 file from which the attributes will be obtained; the default value is "miRNA".
- gffAttribute: the name of the attribute in the GFF3 file to use in the annotations; the default value is "Name".
- qvalue: FDR-corrected pvalue used to filter miRNAs after the differential expression analysis; the default value is "0.05".
- log2FC: Absolute value of the log2FC, used to filter miRNAs after the differential expression analysis; the default value is "0.5".
- distance_method: Method used to compute distances on the hierarchical clustering step; the default value is "euclidean". Availiable methods are: "euclidean", "maximum", "manhattan", "canberra", "binary", "pearson", "abspearson", "correlation", "abscorrelation", "spearman" and "kendall".
A full list of the optional parameters is on the section pipeline parameters.
The conditions_file.txt is a TSV file used by myBrain-Seq to link each fastQ file with its condition. This information will be used to choose the group of samples to compare in the differential expression analysis. Here is an example of a conditions file:
name condition label sex alcohol
C019 control C_before_treatment M 0
C020 control C_before_treatment M 1
C021 control C_after_treatment F 0
C022 control C_after_treatment F 1
P012D FE FE_before_treatment M 1
P013A FE FE_before_treatment F 0
P014A FE FE_after_treatment M 0
P015D FE FE_after_treatment F 1
P014A SEP SEP_before_treatment M 1
P015D SEP SEP_before_treatment F 0
In order to obtain a file with a valid format, the following considerations must be taken into account:
- Columns must be separated by single tabulations.
- The first row must be the header: “name”, “condition” and “label”.
- The first column must be the file rootnames of the fastQ files (i.e.: C019.fastq --> C019).
- The second column must be the conditions.
- The third column is the label, which is only used so that the user can identify each sample in case there is more than one condition. It has no impact on the analysis result but it must be present.
- Additional columns with factors can be included. All these factors will be added to the statistical model of differential expression analysis. Only one factor per column, they can be omitted.
The contrast_file.txt is used by myBrain-Seq to perform the comparisons between samples of two different conditions in the differential expression analysis. Each line on this file corresponds with a contrast that myBrain-Seq has to perform. Here is an example of a contrast file:
name
"Control-First_episode" = "C-FE"
"Control-Second_episode" = "C-SEP"
"First_episode-Second_episode" = "FE-SEP"
The first line of contrast_file.txt is the header, the following lines begin with the contrast label (left side of the equal sign) and the factors to compare (right side of the equal sign). In order to obtain a file with a valid format, the following considerations must be taken into account:
- The first row should be "name", in lowercase.
- The following rows must follow this structure: double quotes, label of the factor to compare, hyphen, label of the reference factor, double quotes, space, equal sign, space, double quotes, factor to compare, hyphen, reference factor, double quotes. No additional spaces should be added, use underscore symbol instead (eg.: First episode should be First_episode). Here is a visual representation of this structure where "B" is the reference factor:
"Label_A-Label_B" = "Factor_A-Factor_B" - The name of the factors to be compared (right side of the equal sign) must be the same as those specified in the "condition" column of the
conditions_file.txt.
Once all the required files were built, to start myBrain-Seq analysis run the script "run.sh" placed on the root of the working directory. This also can be done interactively by using the v.console (see section "Running the v.console"). To run the script manually adapt the following code:
/path/to/working-dir/run.sh /path/to/compi.parametersMyBrain-Seq admits some parameters to customize the execution. Using these parameters you can perform partial executions or control the number of parallel processes of the analysis. These parameters should be quoted and added at the end of the running command:
-
Start myBrain-Seq execution at a specific task:
/path/to/working-dir/run.sh /path/to/compi.parameters "--from task_name" -
Run myBrain-Seq until a specific task:
/path/to/working-dir/run.sh /path/to/compi.parameters "--until task_name" -
Start myBrain-Seq execution after a specific task:
/path/to/working-dir/run.sh /path/to/compi.parameters "--after task_name" -
Change the number of parallel processes (default 5):
/path/to/working-dir/run.sh /path/to/compi.parameters "--num-tasks 2"
You can combine several parameters to gain greater control of the analysis process. Since these parameters are controlled by the Compi framework, please refer to the Compi manual for more information.
Some tasks may produce errors that do not cause the pipeline to fail, but they can be important. Such errors are reported in the log files produced in the logs directory of the pipeline working directory. Inside this directory myBrain-Seq will create additional directories with the logs of each execution, they will be named with the date and hour of the analysis. Files containing the errors are saved with extension *.err.log, whereas normal output is saved with extension *.out.log.
MyBrain-Seq needs the values of some parameters to work, as already indicated in the writing the compi.parameters file section. However, optional parameters with default values can also be edited by adding them to the compi.parameters file. Below is a list of all myBrain-Seq parameters:
| Parameter | Description |
|---|---|
| workingDir | The working directory of the project. |
| fastqDir | The directory containing the fastq files (default is relative to workingDir). |
| outDir | The directory containing the pipeline outputs (relative to workingDir). |
| organism | The organism from which the data was obtained, needed for the functional enrichment analysis. |
| adapter | The sequence of the adapter to remove. |
| genome | The directory path to the genome to align. |
| bwtIndex | The absolute path to the rootname of the Bowtie index. |
| gffFile | The path to the .gff file of the reference genome. |
| gffFeature | Feature of the .gff file to use for the annotations (eg.`: miRNA, gene, transcript...), default miRNA. |
| gffAttribute | Attribute of the .gff to use in the annotations (eg. Name, gene_id, transcript_id...), default "Name". |
| conditions | The path to the .tsv file with the rootnames of the samples, conditions and labels. |
| contrast | The path to the .tsv file with the contrast DESeq2 has to perform. |
| qvalue | FDR-corrected pvalue used to filter miRNAs after the differential expression analysis. |
| log2FC | Absolute value of the log2FC, used to filter miRNAs after the differential expression analysis. |
| distance_method | Method used to compute distances on the hierarchical clustering step, default euclidean. |
| vennFormat | The file format of the Venn diagram (png/svg/tiff), default png. |
| fqcOut | The relative path to the directory containing the FastQC results. |
| ctdOut | The relative path to the directory containing the Cutadapt results. |
| bwtOut | The relative path to the directory containing the Bowtie results. |
| bamstOut | The relative path to the directory containing the Samtools stats and Plot-bamstats results. |
| ftqOut | The relative path to the directory containing the FeatureCounts results. |
| dsqOut | The relative path to the directory containing the DESeq2 results. |
| edgOut | The relative path to the directory containing the EdgeR results. |
| deaIntOut | The relative path to the directory containing the results of the DESeq2 and EdgeR integration. |
| mqcOut | The relative path to the directory containing the MultiQC report. |
| scriptsDir | The relative path to the directory containing the R scripts. |
| testAdapterBashScript | The relative path to the directory containing the R script to get the path of the aligned/unaligned data. |
| deSeq2Rscript | The relative path to the directory containing the R script to run the DESeq2 analysis. |
| filterCtsRscript | The relative path to the directory containing the R script used to filter all-counts.txt and conditions_file. |
| edgerRscript | The relative path to the directory containing the R script to run the EdgeR analysis. |
| enhancedVolcanoRscript | The relative path to the directory containing the R script to build the Volcano plot. |
| deaIntRscript | The relative path to the directory containing the R script to run the DESeq-EdgeR results integration. |
| vennRscript | The relative path to the directory containing the R script to run the DESeq-EdgeR results integration. |
| deseq2NormalizationRscript | The relative path to the directory containing the R script for the creation of the hclust table. |
| hclustMakeTableRscript | The relative path to the directory containing the R script for the creation of the hclust table. |
| hclustRscript | The relative path to the directory containing the R script for the hierarchical clustering analysis. |
| functionalEnrichmentRscript | The relative path to the directory containing the R script for the functional enrichment analysis. |
| networkRscript | The relative path to the directory containing the R script for the network creation. |
| databasesDir | The relative path to the directory containing the TarBase and Reactome databases. |
| tarbaseDB | The relative path to the TarBase file. |
| reactomeDB | The relative path to a Reactome file with Ensembl IDs and Reactome IDs. |
| reactomeInteractionsDB | The relative path to the Reactome file downloaded from here and renamed as ReactomeInteractions.txt. |
| rDeseq2Version | Version of the pegi3s/r_deseq2 Docker image to use. |
| rEdgerVersion | Version of the pegi3s/r_edger Docker image to use. |
| rEnhancedVolcanoVersion | Version of the pegi3s/r_enhanced-volcano Docker image to use. |
| cutadaptVersion | Version of the pegi3s/cutadapt Docker image to use. |
| fastqcVersion | Version of the pegi3s/fastqc Docker image to use. |
| bowtieVersion | Version of the pegi3s/bowtie1 Docker image to use. |
| featureCountsVersion | Version of the pegi3s/feature-counts Docker image to use. |
| samtoolsVersion | Version of the pegi3s/samtools_bcftools Docker image to use. |
| samtoolsBamstatsVersion | Version of the pegi3s/samtools_bcftools Docker image to use for bam analysis. |
| rdatanalysisVersion | Version of the pegi3s/r_data-analysis Docker image to use. |
| rVennVersion | Version of the pegi3s/r_venn-diagram Docker image to use. |
| rNetworkVersion | Version of the pegi3s/r_network Docker image to use. |
| multiqcVersion | Version of the pegi3s/multiqc Docker image to use. |
| selectDEAsoftware | Use this param to select the differential expression analysis software (deseq, edger or both). |
The sample data is available here. Download and decompress it to get a directory named working-dir that contains an example of a functional working directory, were the data and biological references were grouped within it. Here you can find:
- A directory called
input, with thecompi.parameters,condition_file.txtandcontrast_file.txtof this particular study. - A directory called
data, with the fastQ files of the study, the Bowtie index and the miRNA annotations.
To run the pipeline with this test data, edit the compi.parameters (at /working-dir/input) and modify the paths to adapt them to the absolute location of the working directory in your computer (e.g.: workingDir=/working-dir could be workingDir=/home/user/working-dir). After doing this, just run the run.sh script included.
- ≈ 6 minutes - 5 parallel tasks - Ubuntu 20.04.4 LTS, 8 CPUs (Intel® Core™ i7-9700 @ 3.00GHz), 16GB of RAM and SSD disk.
- ≈ 12 minutes - 5 parallel tasks - Ubuntu 18.04.6 LTS, 8 CPUs (Intel® Core™ i7-8565U @ 1.80GHz), 16GB of RAM and SSD disk.
- Pérez-Rodríguez, D., Agís-Balboa, R. C., & López-Fernández, H. (2023). MyBrain-Seq: A Pipeline for MiRNA-Seq Data Analysis in Neuropsychiatric Disorders. Biomedicines, 11(4), Article 4.
- D. Pérez-Rodríguez; M. Pérez-Rodríguez; R.C. Agís-Balboa; H. López-Fernández (2022) Towards a flexible and portable workflow for analyzing miRNA-seq neuropsychiatric data: an initial replicability assessment. 16th International Conference on Practical Applications of Computational Biology & Bioinformatics: PACBB 2022. L'Aquila, Italy. 13 - July
- D. Pérez-Rodríguez; M. Arancha Penedo; T. Rivera-Baltanás; T. Peña-Centeno; S. Burkhardt; A. Fischer; J. M. Prieto-González; J. M. Olivares Díez; H. López-Fernández; R. C. Agís-Balboa (2023) MiRNA differences related to treatment resistant schizophrenia. International Journal of Molecular Sciences. Volume 24(3), 1891. ISSN: 1422-0067
- Pérez-Rodríguez, D., López-Fernández, H., & Agís-Balboa, R. C. (2021). Application of miRNA-seq in neuropsychiatry: A methodological perspective. Computers in Biology and Medicine, 135, 31-42. https://doi.org/10.1016/j.compbiomed.2021.104603
- Pérez-Rodríguez, D., López-Fernández, H., & Agís-Balboa, R. C. (2022). On the Reproducibility of MiRNA-Seq Differential Expression Analyses in Neuropsychiatric Diseases. En M. Rocha, F. Fdez-Riverola, M. S. Mohamad, & R. Casado-Vara (Eds.), Practical Applications of Computational Biology & Bioinformatics, 15th International Conference (PACBB 2021) (pp. 41-51). Springer International Publishing. https://doi.org/10.1007/978-3-030-86258-9_5
To build the Docker image, compi-dk is required. Once you have it installed, simply run compi-dk build -drd -tv from the project directory to build the Docker image. The image will be created with the name specified in the compi.project file. This file also specifies the version of compi that goes into the Docker image.
The pipeline version is set in the <version> section of the pipeline.xml. Nevertheless, as the version number is referenced from other sites, it is recommended to update it using the following command:
./resources/development/set-new-version.sh $(pwd) <new_version>
MyBrain-Seq is a pipeline developed by the SING and NeuroEpigenetics Lab groups.
- Daniel Pérez-Rodríguez
, daniel.prz.rodriguez@gmail.com
- Hugo López-Fernández
, hlfernandez@uvigo.es
- Roberto C. Agís-Balboa
, roberto.carlos.agis.balboa@sergas.es