Deep-learning CNN model for fully automatic glioma segmentation in 3D volume of MRI data
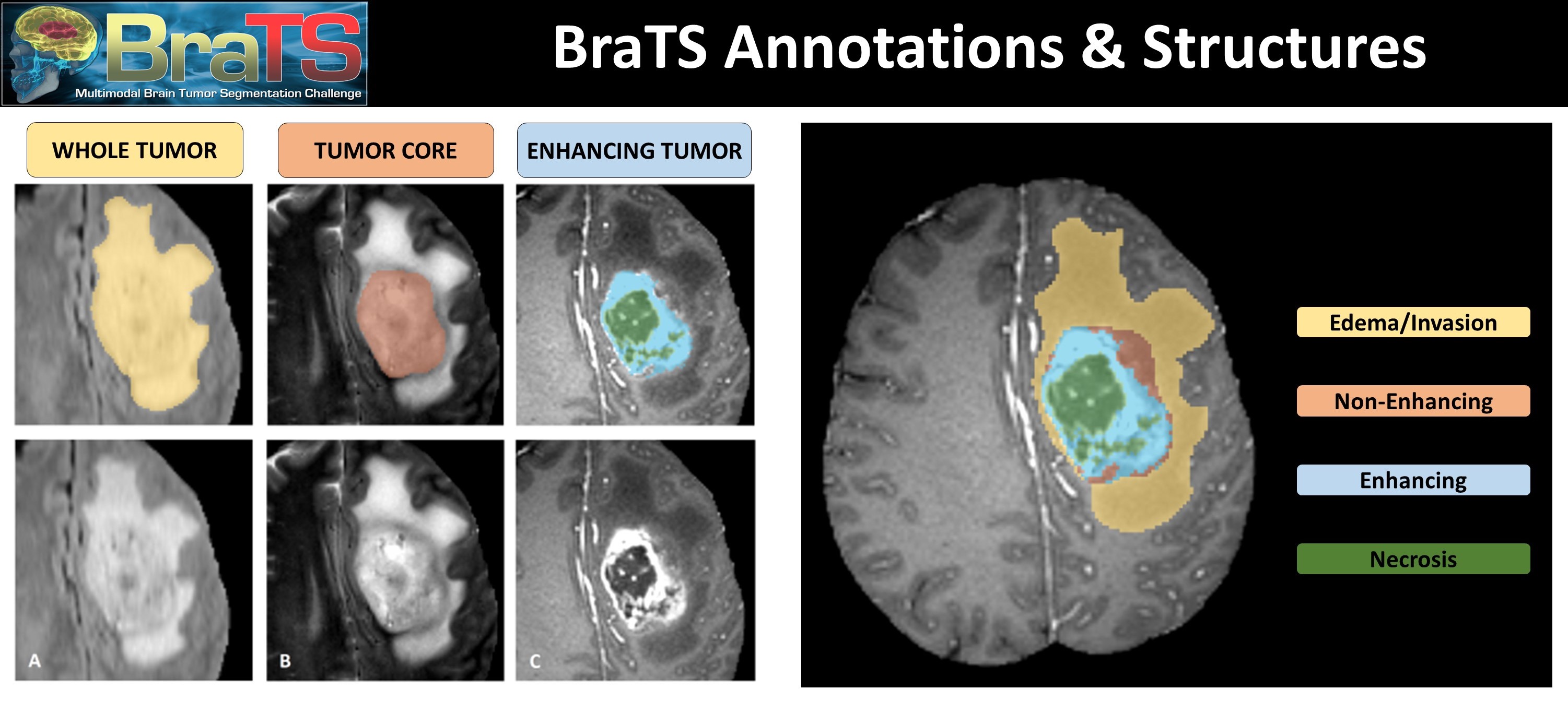
The sub-regions of tumor considered for evaluation are: 1) the "enhancing tumor" (ET), 2) the "tumor core" (TC), and 3) the "whole tumor" (WT) The provided segmentation labels have values of 1 for NCR & NET, 2 for ED, 4 for ET, and 0 for everything else.
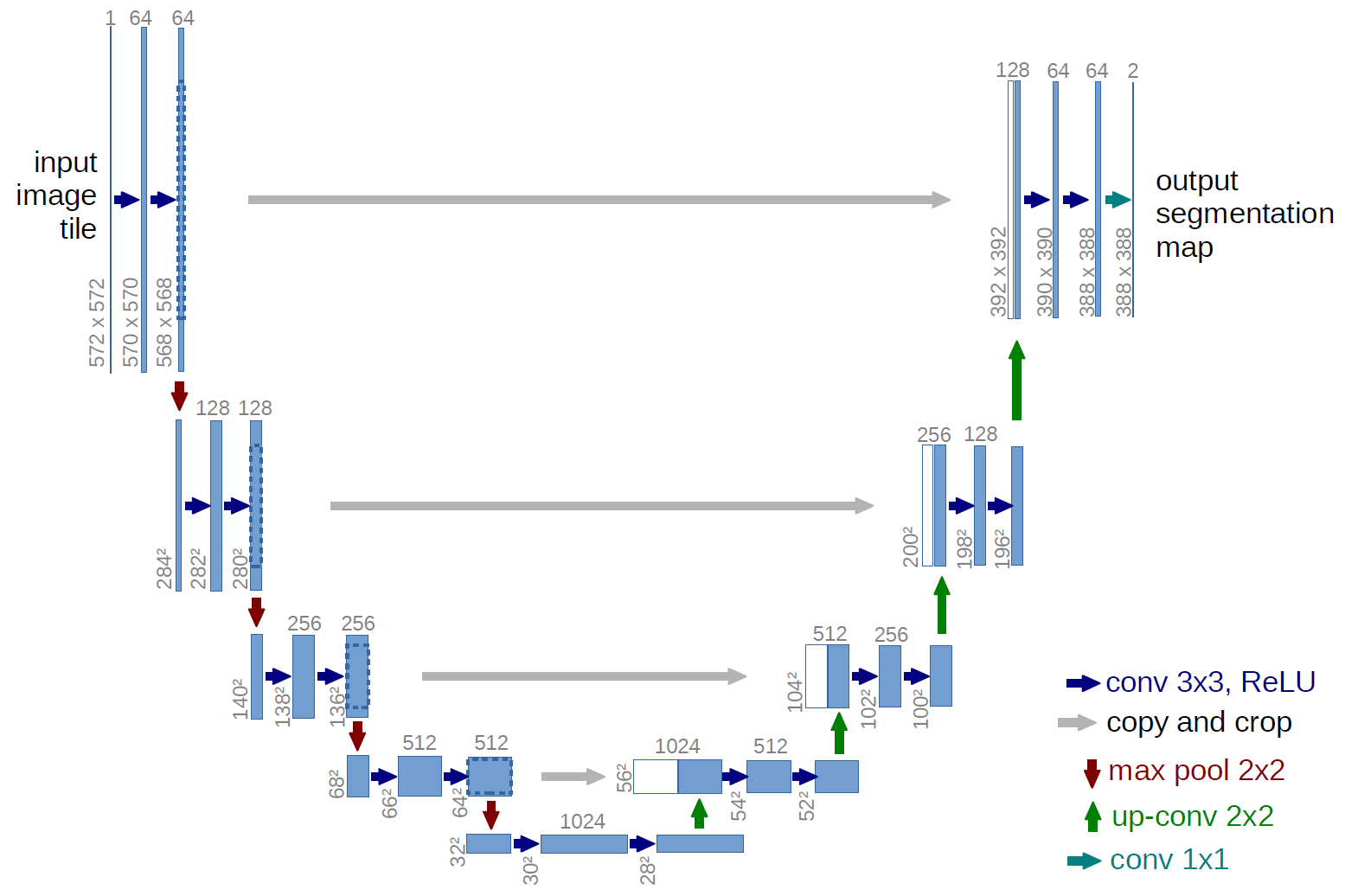
U-NET 2D Architecture as CNN model for tumor segmentation task
U-net is convolutional network architecture for fast and precise segmentation of images. Up to now it has outperformed the prior best method (a sliding-window convolutional network) on the ISBI challenge for segmentation of neuronal structures in electron microscopic stacks.
U-NET kernel initializers: 'he_normal' with 20% propability dropout. Model trained with loss='categorical_crossentropy', Adam optimizer with learning rate 0.001
Mean IoU: 0.8442
Mean dice coeficient: 0.6253
Dice coef class: necrotic: 0.6001
Dice coef class: edema: 0.7287
Dice coef class: enhancing: 0.6366
From left: original slice of a brain, ground truth, all classes predicted, CORE predicted, EDEMA predicted, ENHANCING predicted.
Our task is aimed at classifying patients into three survival groups obtained by unsupervised two-step clustering. These groups roughly correspond to the known survival groups in GBM (PMID: 22517216). The survival groups were characterized as long survivors (e.g., > 450 days), short survivors (e.g., < 300 days), and mid-survivors (e.g., between 300 and 450 days). For precise treatment planning, it is valuable to categorize a patient to either of these survival subgroups. This will enable clinicians to decide how aggressively a patient needs to be treated.
We tried different solutions and compared results. For each model we also implemented GridSearch to find the best parameters for models. For training we are using 4 parameters which are ratios of volumes for each tumor category to the size of a brain and patient's age.
Hyperparameters: {'criterion': 'gini', 'n_estimators': 1}
Train Score: 0.42339181286549704
Test Score: 0.4583333333333333
Hyperparameters: {'C': 10, 'degree': 3, 'gamma': 1, 'kernel': 'poly'}
Train Score: 0.5216374269005847
Test Score: 0.5
Hyperparameters: {'n_neighbors': 38, 'p': 2, 'weights': 'distance'}
Train Score: 0.49941520467836253
Test Score: 0.5416666666666666