Releases: pyg-team/pytorch_geometric
PyG 2.5.3: Bugfixes
PyG 2.5.3 includes a variety of bug fixes related to the MessagePassing refactoring.
Bug Fixes
- Ensure backward compatibility in
MessagePassingviatorch.load(#9105) - Prevent model compilation on custom
propagatefunctions (#9079) - Flush template file before closing it (#9151)
- Do not set
propagatemethod twice inMessagePassingfordecomposed_layers > 1(#9198)
Full Changelog: 2.5.2...2.5.3
PyG 2.5.2: Bugfixes
PyG 2.5.2 includes a bug fix for implementing MessagePassing layers in Google Colab.
Bug Fixes
- Raise error in case
inspect.get_sourceis not supported (#9068)
Full Changelog: 2.5.1...2.5.2
PyG 2.5.1: Bugfixes
PyG 2.5.1 includes a variety of bugfixes.
Bug Fixes
- Ignore
self.propagateappearances in comments when parsingMessagePassingimplementation (#9044) - Fixed
OSErroron read-only file systems withinMessagePassing(#9032) - Made
MessagePassinginterface thread-safe (#9001) - Fixed metaclass conflict in
Dataset(#8999) - Fixed import errors on
MessagePassingmodules with nested inheritance (#8973) - Fix
OSErrorwhen downloading datasets withsimplecache(#8932)
Full Changelog: 2.5.0...2.5.1
PyG 2.5.0: Distributed training, graph tensor representation, RecSys support, native compilation
We are excited to announce the release of PyG 2.5 🎉🎉🎉
PyG 2.5 is the culmination of work from 38 contributors who have worked on features and bug-fixes for a total of over 360 commits since torch-geometric==2.4.0.
Highlights
torch_geometric.distributed
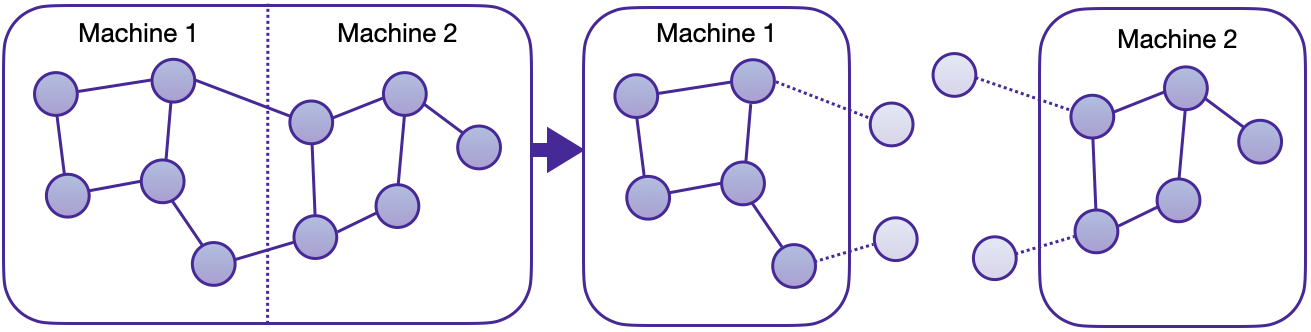
We are thrilled to announce the first in-house distributed training solution for PyG via the torch_geometric.distributed sub-package. Developers and researchers can now take full advantage of distributed training on large-scale datasets which cannot be fully loaded in memory of one machine at the same time. This implementation doesn't require any additional packages to be installed on top of the default PyG stack.
Key Advantages
- Balanced graph partitioning via METIS ensures minimal communication overhead when sampling subgraphs across compute nodes.
- Utilizing DDP for model training in conjunction with RPC for remote sampling and feature fetching routines (with TCP/IP protocol and gloo communication backend) allows for data parallelism with distinct data partitions at each node.
- The implementation via custom
GraphStoreandFeatureStoreAPIs provides a flexible and tailored interface for distributing large graph structure information and feature storage. - Distributed neighbor sampling is capable of sampling in both local and remote partitions through RPC communication channels. All advanced functionality of single-node sampling are also applicable for distributed training, e.g., heterogeneous sampling, link-level sampling, temporal sampling, etc.
- Distributed data loaders offer a high-level abstraction for managing sampler processes, ensuring simplicity and seamless integration with standard PyG data loaders.
See here for the accompanying tutorial. In addition, we provide two distributed examples in examples/distributed/pyg to get started:
- Distributed node-level classification on
ogbn-products - Distributed temporal link prediction on
MovieLens
EdgeIndex Tensor Representation
torch-geometric==2.5.0 introduces the EdgeIndex class.
EdgeIndex is a torch.Tensor, that holds an edge_index representation of shape [2, num_edges]. Edges are given as pairwise source and destination node indices in sparse COO format. While EdgeIndex sub-classes a general torch.Tensor, it can hold additional (meta)data, i.e.:
sparse_size: The underlying sparse matrix sizesort_order: The sort order (if present), either by row or columnis_undirected: Whether edges are bidirectional.
Additionally, EdgeIndex caches data for fast CSR or CSC conversion in case its representation is sorted (i.e. its rowptr or colptr). Caches are filled based on demand (e.g., when calling EdgeIndex.sort_by()), or when explicitly requested via EdgeIndex.fill_cache_(), and are maintained and adjusted over its lifespan (e.g., when calling EdgeIndex.flip()).
from torch_geometric import EdgeIndex
edge_index = EdgeIndex(
[[0, 1, 1, 2],
[1, 0, 2, 1]]
sparse_size=(3, 3),
sort_order='row',
is_undirected=True,
device='cpu',
)
>>> EdgeIndex([[0, 1, 1, 2],
... [1, 0, 2, 1]])
assert edge_index.is_sorted_by_row
assert edge_index.is_undirected
# Flipping order:
edge_index = edge_index.flip(0)
>>> EdgeIndex([[1, 0, 2, 1],
... [0, 1, 1, 2]])
assert edge_index.is_sorted_by_col
assert edge_index.is_undirected
# Filtering:
mask = torch.tensor([True, True, True, False])
edge_index = edge_index[:, mask]
>>> EdgeIndex([[1, 0, 2],
... [0, 1, 1]])
assert edge_index.is_sorted_by_col
assert not edge_index.is_undirected
# Sparse-Dense Matrix Multiplication:
out = edge_index.flip(0) @ torch.randn(3, 16)
assert out.size() == (3, 16)EdgeIndex is implemented through extending torch.Tensor via the __torch_function__ interface (see here for the highly recommended tutorial).
EdgeIndex ensures for optimal computation in GNN message passing schemes, while preserving the ease-of-use of regular COO-based PyG workflows. EdgeIndex will fully deprecate the usage of SparseTensor from torch-sparse in later releases, leaving us with just a single source of truth for representing graph structure information in PyG.
RecSys Support
Previously, all/most of our link prediction models were trained and evaluated using binary classification metrics. However, this usually requires that we have a set of candidates in advance, from which we can then infer the existence of links. This is not necessarily practical, since in most cases, we want to find the top-k most likely links from the full set of O(N^2) pairs.
torch-geometric==2.5.0 brings full support for using GNNs as a recommender system (#8452), including support for
- Maximum Inner Product Search (MIPS) via
MIPSKNNIndex - Retrieval metrics such as
f1@k,map@k,precision@k,recall@kandndcg@k, including mini-batch support
mips = MIPSKNNIndex(dst_emb)
for src_batch in src_loader:
src_emb = model(src_batch.x_dict, src_batch.edge_index_dict)
_, pred_index_mat = mips.search(src_emb, k)
for metric in retrieval_metrics:
metric.update(pred_index_mat, edge_label_index)
for metric in retrieval_metrics:
metric.compute()See here for the accompanying example.
PyTorch 2.2 Support
PyG 2.5 is fully compatible with PyTorch 2.2 (#8857), and supports the following combinations:
| PyTorch 2.2 | cpu |
cu118 |
cu121 |
|---|---|---|---|
| Linux | ✅ | ✅ | ✅ |
| macOS | ✅ | ||
| Windows | ✅ | ✅ | ✅ |
You can still install PyG 2.5 with an older PyTorch release up to PyTorch 1.12 in case you are not eager to update your PyTorch version.
Native torch.compile(...) and TorchScript Support
torch-geometric==2.5.0 introduces a full re-implementation of the MessagePassing interface, which makes it natively applicable to both torch.compile and TorchScript. As such, torch_geometric.compile is now fully deprecated in favor of [torch.compile](https://pytorch.org/docs/stable/generated/tor...
PyG 2.4.0: Model compilation, on-disk datasets, hierarchical sampling
We are excited to announce the release of PyG 2.4 🎉🎉🎉
PyG 2.4 is the culmination of work from 62 contributors who have worked on features and bug-fixes for a total of over 500 commits since torch-geometric==2.3.1.
Highlights
PyTorch 2.1 and torch.compile(dynamic=True) support
The long wait has an end! With the release of PyTorch 2.1, PyG 2.4 now brings full support for torch.compile to graphs of varying size via the dynamic=True option, which is especially useful for use-cases that involve the usage of DataLoader or NeighborLoader. Examples and tutorials have been updated to reflect this support accordingly (#8134), and models and layers in torch_geometric.nn have been tested to produce zero graph breaks:
import torch_geometric
model = torch_geometric.compile(model, dynamic=True)When enabling the dynamic=True option, PyTorch will up-front attempt to generate a kernel that is as dynamic as possible to avoid recompilations when sizes change across mini-batches changes. As such, you should only ever not specify dynamic=True when graph sizes are guaranteed to never change. Note that dynamic=True requires PyTorch >= 2.1.0 to be installed.
PyG 2.4 is fully compatible with PyTorch 2.1, and supports the following combinations:
| PyTorch 2.1 | cpu |
cu118 |
cu121 |
|---|---|---|---|
| Linux | ✅ | ✅ | ✅ |
| macOS | ✅ | ||
| Windows | ✅ | ✅ | ✅ |
You can still install PyG 2.4 on older PyTorch releases up to PyTorch 1.11 in case you are not eager to update your PyTorch version.
OnDiskDataset Interface
We added the OnDiskDataset base class for creating large graph datasets (e.g., molecular databases with billions of graphs), which do not easily fit into CPU memory at once (#8028, #8044, #8046, #8051, #8052, #8054, #8057, #8058, #8066, #8088, #8092, #8106). OnDiskDataset leverages our newly introduced Database backend (sqlite3 by default) for on-disk storage and access of graphs, supports DataLoader out-of-the-box, and is optimized for maximum performance.
OnDiskDataset utilizes a user-specified schema to store data as efficient as possible (instead of Python pickling). The schema can take int, float str, object or a dictionary with dtype and size keys (for specifying tensor data) as input, and can be nested as a dictionary. For example,
dataset = OnDiskDataset(root, schema={
'x': dict(dtype=torch.float, size=(-1, 16)),
'edge_index': dict(dtype=torch.long, size=(2, -1)),
'y': float,
})creates a database with three columns, where x and edge_index are stored as binary data, and y is stored as a float.
Afterwards, you can append data to the OnDiskDataset and retrieve data from it via dataset.append()/dataset.extend(), and dataset.get()/dataset.multi_get(), respectively. We added a fully working example on how to set up your own OnDiskDataset here (#8102). You can also convert in-memory dataset instances to an OnDiskDataset instance by running InMemoryDataset.to_on_disk_dataset() (#8116).
Neighbor Sampling Improvements
Hierarchical Sampling
One drawback of NeighborLoader is that it computes a representations for all sampled nodes at all depths of the network. However, nodes sampled in later hops no longer contribute to the node representations of seed nodes in later GNN layers, thus performing useless computation. NeighborLoader will be marginally slower since we are computing node embeddings for nodes we no longer need. This is a trade-off we have made to obtain a clean, modular and experimental-friendly GNN design, which does not tie the definition of the model to its utilized data loader routine.
With PyG 2.4, we introduced the option to eliminate this overhead and speed-up training and inference in mini-batch GNNs further, which we call "Hierarchical Neighborhood Sampling" (see here for the full tutorial) (#6661, #7089, #7244, #7425, #7594, #7942). Its main idea is to progressively trim the adjacency matrix of the returned subgraph before inputting it to each GNN layer, and works seamlessly across several models, both in the homogeneous and heterogeneous graph setting. To support this trimming and implement it effectively, the NeighborLoader implementation in PyG and in pyg-lib additionally return the number of nodes and edges sampled in each hop, which are then used on a per-layer basis to trim the adjacency matrix and the various feature matrices to only maintain the required amount (see the trim_to_layer method):
class GNN(torch.nn.Module):
def __init__(self, in_channels: int, out_channels: int, num_layers: int):
super().__init__()
self.convs = ModuleList([SAGEConv(in_channels, 64)])
for _ in range(num_layers - 1):
self.convs.append(SAGEConv(hidden_channels, hidden_channels))
self.lin = Linear(hidden_channels, out_channels)
def forward(
self,
x: Tensor,
edge_index: Tensor,
num_sampled_nodes_per_hop: List[int],
num_sampled_edges_per_hop: List[int],
) -> Tensor:
for i, conv in enumerate(self.convs):
# Trim edge and node information to the current layer `i`.
x, edge_index, _ = trim_to_layer(
i, num_sampled_nodes_per_hop, num_sampled_edges_per_hop,
x, edge_index)
x = conv(x, edge_index).relu()
return self.lin(x)Corresponding examples can be found here and here.
Biased Sampling
Additionally, we added support for weighted/biased sampling in NeighborLoader/LinkNeighborLoader scenarios. For this, simply specify your edge_weight attribute during NeighborLoader initialization, and PyG will pick up these weights to perform weighted/biased sampling (#8038):
data = Data(num_nodes=5, edge_index=edge_index, edge_weight=edge_weight)
loader = NeighborLoader(
data,
num_neighbors=[10, 10],
weight_attr='edge_weight',
)
batch = next(iter(loader))New models, datasets, examples & tutorials
As part of our algorithm and documentation sprints (#7892), we have added:
- **Model components:*...
Pyg 2.3.1: Bugfixes
PyG 2.3.1 includes a variety of bugfixes.
Bug Fixes
- Fixed
cugraphGNN layer support forpylibcugraphops==23.04(#7023) - Removed
DeprecationWarningofTypedStorageusage inDataLoader(#7034) - Fixed a bug in
FastHGTConvthat computed values via parameters used to compute the keys (#7050) - Fixed
numpyincompatiblity when reading files inPlanetoiddatasets (#7141) - Fixed
utils.subgraphon unordered inputs (#7187) - Fixed support for
Data.num_edgesfor nativetorch.sparse.Tensoradjacency matrices (#7104)
Full Changelog: 2.3.0...2.3.1
Pyg 2.3.0: PyTorch 2.0 support, native sparse tensor support, explainability and accelerations
We are thrilled to announce the release of PyG 2.3 🎉
PyG 2.3 is the culmination of work from 59 contributors who have worked on features and bug-fixes for a total of over 470 commits since torch-geometric==2.2.0.
Highlights
PyTorch 2.0 Support
PyG 2.3 is fully compatible with the next generation release of PyTorch, bringing many new innovations and features such as torch.compile() and Python 3.11 support to PyG out-of-the-box. In particular, many PyG models and functions are speeded up significantly using torch.compile() in torch >= 2.0.0.
We have prepared a full tutorial and a set of examples to get you going with torch.compile() immediately:
import torch_geometric
from torch_geometric.nn import GraphSAGE
model = GraphSAGE(in_channels, hidden_channels, num_layers, out_channels)
model = model.to(device)
model = torch_geometric.compile(model)Overall, we observed runtime improvements of nearly up to 300%:
| Model | Mode | Forward | Backward | Total | Speedup |
|---|---|---|---|---|---|
GCN |
Eager | 2.6396s | 2.1697s | 4.8093s | |
GCN |
Compiled | 1.1082s | 0.5896s | 1.6978s | 2.83x |
GraphSAGE |
Eager | 1.6023s | 1.6428s | 3.2451s | |
GraphSAGE |
Compiled | 0.7033s | 0.7465s | 1.4498s | 2.24x |
GIN |
Eager | 1.6701s | 1.6990s | 3.3690s | |
GIN |
Compiled | 0.7320s | 0.7407s | 1.4727s | 2.29x |
Please note that torch.compile() within PyG is in beta mode and under active development. For example, currently torch.compile(model, dynamic=True) does not yet work seamlessly, but fixes are on its way. We are very eager to improve its support across the whole PyG code base, so do not hesitate to reach out if you notice anything unexpected.
Infrastructure Changes
With the recent upstreams of torch-scatter and torch-sparse to native PyTorch, we are happy to announce that any installation of the extension packages torch-scatter, torch-sparse, torch-cluster and torch-spline-conv is now fully optional.
All it takes to install PyG is now encapsulated into a single command
pip install torch-geometric
and finally resolves a lot of previous installation issues.
Extension packages are still picked up for the following use-cases (if installed):
pyg-lib: Heterogeneous GNN operators and graph sampling routines likeNeighborLoadertorch-scatter: Accelerated"min"and"max"reductionstorch-sparse:SparseTensorsupporttorch-cluster: Graph clustering routines likeknnorradiustorch-spline-conv:SplineConvsupport
We recommend to start with a minimal installation, and only install additional dependencies once you actually get notified about them being missing during PyG usage.
Native PyTorch Sparse Tensor Support
With the recent additions of torch.sparse_csr_tensor and torch.sparse_csc_tensor classes and accelerated sparse matrix multiplication routines to PyTorch, we finally enable MessagePassing on pure PyTorch sparse tensors as well. In particular, you can now use torch.sparse_csr_tensor and torch.sparse_csc_tensor as a drop-in replacement for torch_sparse.SparseTensor:
from torch_geometric.nn import GCN
import torch_geometric.transforms as T
from torch_geometric.datasets import Planetoid
transform = T.ToSparseTensor(layout=torch.sparse_csr)
dataset = Planetoid("Planetoid", name="Cora", transform=transform)
model = GCN(in_channels, hidden_channels, num_layers=2)
model = model(data.x, data.adj_t)Nearly all of the native PyG layers have been tested to work seamlessly with native PyTorch sparse tensors (#5906, #5944, #6003, #6033, #6514, #6532, #6748, #6847, #6868, #6874, #6897, #6930, #6932, #6936, #6937, #6939, #6947, #6950, #6951, #6957).
Explainability Framework
In PyG 2.2 we introduced the torch_geometric.explain package that provides a flexible interface to generate and visualize GNN explanations using various algorithms. We are happy to add the following key improvements on this front:
- Support for explaining heterogeneous GNNs via
HeteroExplanation - New visualization tools to
visualize_feature_importanceand tovisualize_graphexplanations - May new datasets and metrics to evaluate explanation algorithms
- Several new explanation algorithms such as
CaptumExplainer,PGExplainer,AttentionExplainer,PGMExplainer, andGraphMaskExplainer - Support for node-level, link-level and graph-level explanations
Using the new explainer interface is as simple as:
explainer = Explainer(
model=model,
algorithm=CaptumExplainer('IntegratedGradients'),
explanation_type='model',
model_config=dict(
mode='multiclass_classification',
task_level='node',
return_type='log_probs',
),
node_mask_type='attributes',
edge_mask_type='object',
)
explanation = explainer(data.x, data.edge_index)Read more about torch_geometric.explain in our newly added tutorial and example scripts. We also added a blog post that describes the new int...
PyG 2.2.0: Accelerations and Scalability
We are excited to announce the release of PyG 2.2 🎉🎉🎉
PyG 2.2 is the culmination of work from 78 contributors who have worked on features and bug-fixes for a total of over 320 commits since torch-geometric==2.1.0.
Highlights
pyg-lib Integration
We are proud to release and integrate pyg-lib==0.1.0 into PyG, the first stable version of our new low-level Graph Neural Network library to drive all CPU and GPU acceleration needs of PyG (#5330, #5347, #5384, #5388).
You can install pyg-lib as described in our README.md:
pip install pyg-lib -f https://data.pyg.org/whl/torch-${TORCH}+${CUDA}.html
import pyg_libOnce pyg-lib is installed, it will get automatically picked up by PyG, e.g., to accelerate neighborhood sampling routines or to accelerate heterogeneous GNN execution:
pyg-libprovides fast and optimized CPU routines to iteratively sample neighbors in homogeneous and heterogeneous graphs, and heavily improves upon the previously used neighborhood sampling techniques utilized in PyG.
pyg-libprovides efficient GPU-based routines to parallelize workloads in heterogeneous graphs across different node types and edge types. We achieve this by leveraging type-dependent transformations via NVIDIA CUTLASS integration, which is flexible to implement most heterogeneous GNNs with, and efficient, even for sparse edge types or a large number of different node types.
GraphStore and FeatureStore Abstractions
PyG 2.2 includes numerous primitives to easily integrate with simple paradigms for scalable graph machine learning, enabling users to train GNNs on graphs far larger than the size of their machine's available memory. It does so by introducing simple, easy-to-use, and extensible abstractions of a FeatureStore and a GraphStore that plug directly into existing familiar PyG interfaces (see here for the accompanying tutorial).
feature_store = CustomFeatureStore()
feature_store['paper', 'x', None] = ... # Add paper features
feature_store['author', 'x', None] = ... # Add author features
graph_store = CustomGraphStore()
graph_store['edge', 'coo'] = ... # Add edges in "COO" format
# `CustomGraphSampler` knows how to sample on `CustomGraphStore`:
graph_sampler = CustomGraphSampler(
graph_store=graph_store,
num_neighbors=[10, 20],
...
)
from torch_geometric.loader import NodeLoader
loader = NodeLoader(
data=(feature_store, graph_store),
node_sampler=graph_sampler,
batch_size=20,
input_nodes='paper',
)
for batch in loader:
passData loading and sampling routines are refactored and decomposed into torch_geometric.loader and torch_geometric.sampler modules, respectively (#5563, #5820, #5456, #5457, #5312, #5365, #5402, #5404, #5418).
Optimized and Fused Aggregations
PyG 2.2 further accelerates scatter aggregations based on CPU/GPU and with/without backward computation paths (requires torch>=1.12.0 and torch-scatter>=2.1.0) (#5232, #5241, #5353, #5386, #5399, #6051, #6052).
We also optimized the usage of nn.aggr.MultiAggregation by fusing the computation of multiple aggregations together (see here for more details) (#6036, #6040).
Here are some benchmarking results on PyTorch 1.12 (summed over 1000 runs):
| Aggregators | Vanilla | Fusion |
|---|---|---|
[sum, mean] |
0.3325s | 0.1996s |
[sum, mean, min, max] |
0.7139s | 0.5037s |
[sum, mean, var] |
0.6849s | 0.3871s |
[sum, mean, var, std] |
1.0955s | 0.3973s |
Lastly, we have incorporated "fused" GNN operators via the dgNN package, starting with a FusedGATConv implementation (#5140).
Community Sprint: Type Hints and TorchScript Support
We are running regular community sprints to get our community more involved in building PyG. Whether you are just beginning to use graph learning or have been leveraging GNNs in research or production, the community sprints welcome members of all levels with different types of projects.
We had our first community sprint on 10/12 to fully-incorporate type hints and TorchScript support over the entire code base. The goal was to improve usability and cleanliness of our codebase. We had 20 contributors participating, contributing to 120 type hints within 2 weeks, adding around 2400 lines of code (#5842, #5603, #5659, #5664, #5665, #5666, #5667, #5668, #5669, #5673, #5675, #5673, #5678, #5682, #5683, #5684, #5685, #5687, #5688, #5695, #5699, #5701, #5702, #5703, #5706, #5707, #5710, #5714, #5715, #5716, #5722, #5724, #5725, #5726, #5729, #5730, #5731, #5732, [#5733](https://github.com/pyg-team/pyt...
PyG 2.1.0: Principled aggregations, link-level and temporal samplers, data pipe support, ...
We are excited to announce the release of PyG 2.1.0 🎉🎉🎉
PyG 2.1.0 is the culmination of work from over 60 contributors who have worked on features and bug-fixes for a total of over 320 commits since torch-geometric==2.0.4.
Highlights
Principled Aggregations
See here for the accompanying tutorial.
Aggregation functions play an important role in the message passing framework and the readout functions of Graph Neural Networks. Specifically, many works in the literature (Hamilton et al. (2017), Xu et al. (2018), Corso et al. (2020), Li et al. (2020), Tailor et al. (2021), Bartunov et al. (2022)) demonstrate that the choice of aggregation functions contributes significantly to the representational power and performance of the model.
To facilitate further experimentation and unify the concepts of aggregation within GNNs across both MessagePassing and global readouts, we have made the concept of Aggregation a first-class principle in PyG (#4379, #4522, #4687, #4721, #4731, #4762, #4749, #4779, #4863, #4864, #4865, #4866, #4872, #4927, #4934, #4935, #4957, #4973, #4973, #4986, #4995, #5000, #5021, #5034, #5036, #5039, #4522, #5033, #5085, #5097, #5099, #5104, #5113, #5130, #5098, #5191). As of now, PyG provides support for various aggregations — from simple ones (e.g., mean, max, sum), to advanced ones (e.g., median, var, std), learnable ones (e.g., SoftmaxAggregation, PowerMeanAggregation), and exotic ones (e.g., LSTMAggregation, SortAggregation, EquilibriumAggregation). Furthermore, multiple aggregations can be combined and stacked together:
from torch_geometric.nn import MessagePassing, SoftmaxAggregation
class MyConv(MessagePassing):
def __init__(self, ...):
# Combines a set of aggregations and concatenates their results.
# The interface also supports automatic resolution.
super().__init__(aggr=['mean', 'std', SoftmaxAggregation(learn=True)])Link-level Neighbor Loader
We added a new LinkNeighborLoader class for training scalable GNNs that perform edge-level predictions on giant graphs (#4396, #4439, #4441, #4446, #4508, #4509, #4868). LinkNeighborLoader comes with automatic support for both homogeneous and heterogenous data, and supports link prediction via automatic negative sampling as well as edge-level classification and regression models:
from torch_geometric.loader import LinkNeighborLoader
loader = LinkNeighborLoader(
data,
num_neighbors=[30] * 2, # Sample 30 neighbors for each node for 2 iterations
batch_size=128, # Use a batch size of 128 for sampling training links
edge_label_index=data.edge_index, # Use the entire graph for supervision
negative_sampling_ratio=1.0, # Sample negative edges
)
sampled_data = next(iter(loader))
print(sampled_data)
>>> Data(x=[1368, 1433], edge_index=[2, 3103], edge_label_index=[2, 256], edge_label=[256])Neighborhood Sampling based on Temporal Constraints
Both NeighborLoader and LinkNeighborLoader now support temporal sampling via the time_attr argument (#4025, #4877, #4908, #5137, #5173). If set, temporal sampling will be used such that neighbors are guaranteed to fulfill temporal constraints, i.e. neighbors have an earlier timestamp than the center node:
from torch_geometric.loader import NeighborLoader
data['paper'].time = torch.arange(data['paper'].num_nodes)
loader = NeighborLoader(
data,
input_nodes='paper',
time_attr='time', # Only sample papers that appeared before the seed paper
num_neighbors=[30] * 2,
batch_size=128,
)Note that this feature requires torch-sparse>=0.6.14.
Functional DataPipes
See here for the accompanying example.
PyG now fully supports data loading using the newly introduced concept of DataPipes in PyTorch for easily constructing flexible and performant data pipelines (#4302, #4345, #4349). PyG provides DataPipe support for batching multiple PyG data objects together and for applying any PyG transform:
datapipe = FileOpener(['SMILES_HIV.csv'])
datapipe = datapipe.parse_csv_as_dict()
datapipe = datapipe.parse_smiles(target_key='HIV_active')
datapipe = datapipe.in_memory_cache() # Cache graph instances in-memory.
datapipe = datapipe.shuffle()
datapipe = datapipe.batch_graphs(batch_size=32)datapipe = FileLister([root_dir], masks='*.off', recursive=True)
datapipe = datapipe.read_mesh()
datapipe = datapipe.in_memory_cache() # Cache graph instances in-memory.
datapipe = datapipe.sample_points(1024) # Use PyG transforms from here.
datapipe = datapipe.knn_graph(k=8)
datapipe = datapipe.shuffle()
datapipe = datapipe.batch_graphs(batch_size=32)Breaking Changes
- The
torch_geometric.utils.metricpackage has been removed. We now recommend to use thetorchmetricspacka...
2.0.4
PyG 2.0.4 🎉
A new minor PyG version release, bringing PyTorch 1.11 support to PyG. It further includes a variety of new features and bugfixes:
Features
- Added Quiver examples for multi-GU training using
GraphSAGE(#4103), thanks to @eedalong and @luomai nn.model.to_captum: Full integration of explainability methods provided by the Captum library (#3990, #4076), thanks to @RBendiasnn.conv.RGATConv: The relational graph attentional operator (#4031, #4110), thanks to @fork123aniketnn.pool.DMoNPooling: The spectral modularity pooling operator (#4166, #4242), thanks to @fork123aniketnn.*: Support for shape information in the documentation (#3739, #3889, #3893, #3946, #3981, #4009, #4120, #4158), thanks to @saiden89 and @arunppsg and @konstantinosKokosloader.TemporalDataLoader: A dataloader to load aTemporalDataobject in mini-batches (#3985, #3988), thanks to @otaviocxloader.ImbalancedSampler: A weighted random sampler that randomly samples elements according to class distribution (#4198)transforms.VirtualNode: A transform that adds a virtual node to a graph (#4163)transforms.LargestConnectedComponents: Selects the subgraph that corresponds to the largest connected components in the graph (#3949), thanks to @abojchevskiutils.homophily: Support for class-insensitive edge homophily (#3977, #4152), thanks to @hash-ir and @jinjh0123utils.get_mesh_laplacian: Mesh Laplacian computation (#4187), thanks to @daniel-unyi-42
Datasets
- Added a dataset cheatsheet to the documentation that collects import graph statistics across a variety of datasets supported in PyG (#3807, #3817) (please consider helping us filling its remaining content)
datasets.EllipticBitcoinDataset: A dataset of Bitcoin transactions (#3815), thanks to @shravankumar147
Minor Changes
nn.models.MLP: MLPs can now either be initialized via a list ofchannelsor by specifyinghidden_channelsandnum_layers(#3957)nn.models.BasicGNN: FinalLineartransformations are now always applied (except forjk=None) (#4042)nn.conv.MessagePassing: Message passing modules that make use ofedge_updaterare now jittable (#3765), thanks to @Padarnnn.conv.MessagePassing: (Official) support forminandmulaggregations (#4219)nn.LightGCN: Initialize embeddings viaxavier_uniformfor better model performance (#4083), thanks to @nishithshowri006nn.conv.ChebConv: Automatic eigenvalue approximation (#4106), thanks to @daniel-unyi-42nn.conv.APPNP: Added support for optionaledge_weight, (690a01d), thanks to @YueeXiangnn.conv.GravNetConv: Support fortorch.jit.script(#3885), thanks to @RobMcHnn.pool.global_*_pool: Thebatchvector is now optional (#4161)nn.to_hetero: Added a warning in caseto_heterois used onHeteroDatametadata with unused destination node types (#3775)nn.to_hetero: Support for nested modules (ea135bf)nn.Sequential: Support for indexing (#3790)nn.Sequential: Support forOrderedDictas input (#4075)datasets.ZINC: Added an in-depth description of the task (#3832), thanks to @gasteigerjodatasets.FakeDataset: Support for different feature distributions across different labels (#4065), thanks to @arunppsgdatasets.FakeDataset: Support for custom global attributes (#4074), thanks to @arunppsgtransforms.NormalizeFeatures: Features will no longer be transformed in-place (ada5b9a)transforms.NormalizeFeatures: Support for negative feature values (6008e30)utils.is_undirected: Improved efficiency (#3789)utils.dropout_adj: Improved efficiency (#4059)utils.contains_isolated_nodes: Improved efficiency (970de13)utils.to_networkx: Support forto_undirectedoptions (upper triangle vs. lower triangle) (#3901, #3948), thanks to @RemyLaugraphgym: Support for custom metrics and loggers (#3494), thanks to @RemyLaugraphgym.register: Register operations can now be used as class decorators (#3779, #3782)- Documentation: Added a few exercises at the end of documentation tutorials (#3780), thanks to @PabloAMC
- Documentation: Added better installation instructions to
CONTRIBUTUNG.md(#3803, #3991, #3995), thanks to @Cho-Geonwoo and @RBendias and @RodrigoVillatoro - Refactor: Clean-up dependencies (#3908, #4133, #4172), thanks to @adelizer
- CI: Improved test runtimes (#4241)
- CI: Additional linting check via
yamllint(#3886) - CI: Additional linting check via
isort(66b1780), thanks to @mananshah99 torch.package: Model packaging viatorch.package(#3997)
Bugfixes
data.HeteroData: Fixed a bug indata.{attr_name}_dictin casedata.{attr_name}does not exist (#3897)data.Data: Fixeddata.is_edge_attrin casedata.num_edges == 1(#3880)data.Batch: Fixed a device mismatch bug in case abatchobject was indexed that was created from GPU tensors (e6aa4c9, c549b3b)
*...