Jing Ai and Jung Hoon Son
BINF G4002 Computation Methods
Final Project
**Utilizing TCRD database (used by Pharos) to predict drug targets associated with adverse drug events. **
R workspace file, containing all_tcrd data frame for our analysis. This data frame can be generated from Step1_SQL_query_matrix_builder.R file on any computer with a local instance of Pharos/TCRD database installed on MySQL. SQL data dump is available from: (http://juniper.health.unm.edu/tcrd/download/tcrd_v4.4.2.sql.gz)
Individual IDG family based data sets (GPCR, Ion channels, Kinases, NRs, null-classes) have been compiled and is made available in the folder individual_class.
toxic_targets/GoldStandards.csv -This file contains curated information from WITHDRAWN database, implicating drug targets associated with withdrawn drugs around the world (rows where y=0). Together with the protein targets classified as Tclin in TCRD that are not associated with any withdrawn drugs (rows where y=1), they serves as our gold standards for denoting which targets have been implicated with drug adverse events and which have not.
Step1_SQL_query_matrix_builder.R
This code queries each druggable target + additional targets identified via WITHDRAWN database from a local instance of TCRD MySQL database. It expands the feature set with each new target vector representation added.
analysis_code/DataMappingColsums.R
The file contains code for feature reduction. Since a large number of features contained zeros, we first removed the columns with less than 20 non-zero values (out of 1129 samples).
analysis_code/FeatureSelection.ipynb
This file contains code for feature selection codes (in python). We applied chi-square feature selection and filered out features with chi-square p-values of <0.05
Step2_prelim_prediction.R is used for preliminary randomForest model in R, for quickly testing our test sets.
analysis_code/combined_analysis_JA.R The files contains the modeling codes for L1-Logistic Regression (5-fold CV), Naive Bayes, Random Forrest based on cross validation.
analysis_code/SVM.R The file contains the modeling code for Support Vector Machine (Linear and Kernel).
future_work/predicting_drugs.R This code can be used to generate an adverse drug scoring system. We query the entire TCRD database for drugs associated with targets (among the 2117 we predicted) predicted by our algorithm. We obtain a score using
Drug_prediction_score.csv is generated by compiling all targets predicted by a exploratory randomForest model, and compiling the following score:
Score =
# of Predicted Adverse Targets-# of Predicted non-Adverse targets.
In which drugs with higest score we expect to have highest likelihood of being implicated with an adverse drug effect.
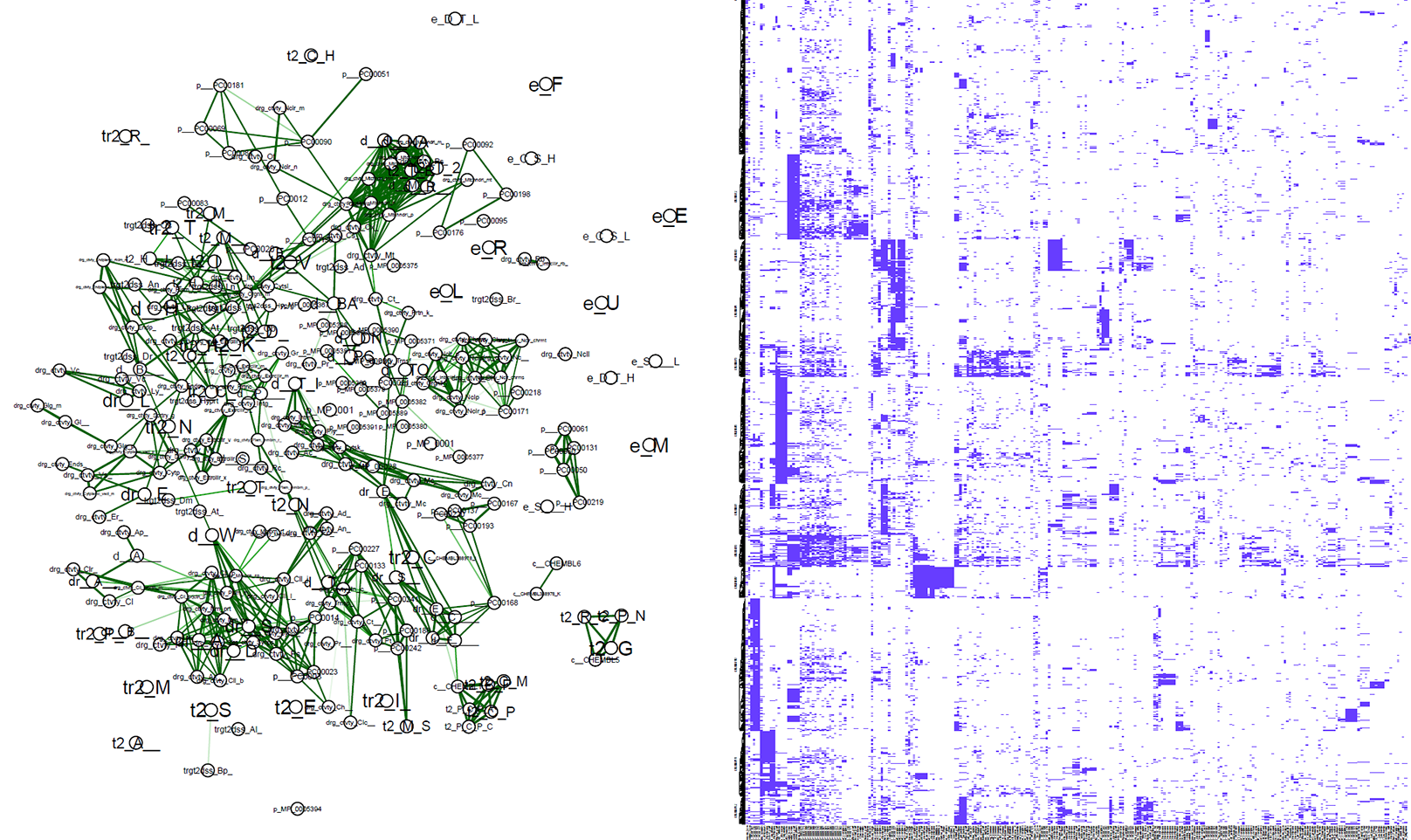
future_work/plot_playground.R This is an experimental file, used to generate exploratory heatmaps and network visualizations. It currently generates two plots:
heatmap.pdf is a demonstration of heatmaps with K-means clustering applied, showing similar targets based on our vector representation.
correlation_network.pdf is a visualization of correlation matrix of the features. Uninformative, but shows potential.
This folder contains our results and model performance and evaluations. Important feature lists are contained within this folder.