This repository contains biomedical discourse treebanks:
- COVID19-DTB (Nishida and Matsumoto, 2021; to appear in TACL)
COVID19-DTB is a collection of manually annotated discourse dependency structures for scholarly paper abstracts on COVID-19 and related coronaviruses like SARS and MERS.
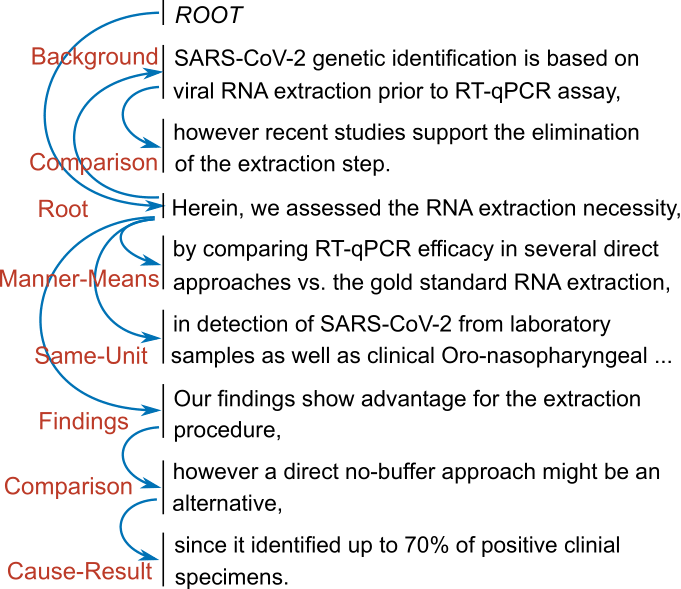
The following figure shows an actual example of discourse dependency structure for an abstract (Israeli et al., 2020) in COVID19-DTB.
First, we sampled 300 abstracts randomly from the 2020 September snapshot of The COVID-19 Open Research Dataset (CORD-19). Then, the 300 abstracts were segmented into Elementary Discourse Units (EDUs) manually by the authors. Then, we employed two professional annotators to give gold discourse dependency structures to the 300 abstracts independently. We divided the results into development and test sets, each of which consists of 150 examples.
The further details on the annotation scheme, annotation procedure, and corpus statistics can be found in our paper.
We follow the same JSON schema with the SciDTB dataset (Yang and Li, 2018): Each discourse dependency structure is stored as a JSON file like the following:
{
"root": [
{
"id": 0,
"parent": -1,
"text": "ROOT",
"relation": "null"
},
{
"id": 1,
"parent": 3,
"text": "SARS - CoV-2 genetic identification is based on viral RNA extraction prior to RT - qPCR assay ,",
"relation": "BACKGROUND"
},
{
"id": 2,
"parent": 1,
"text": "however recent studies support the elimination of the extraction step . <S>",
"relation": "COMPARISON"
},
{
"id": 3,
"parent": 0,
"text": "Herein , we assessed the RNA extraction necessity ,",
"relation": "ROOT"
},
{
"id": 4,
"parent": 3,
"text": "by comparing RT - qPCR efficacy in several direct approaches vs. the gold standard RNA extraction ,",
"relation": "MANNER-MEANS"
},
{
"id": 5,
"parent": 3,
"text": "in detection of SARS - CoV-2 from laboratory samples as well as clinical Oro - nasopharyngeal SARS - CoV-2 swabs . <S>",
"relation": "SAME-UNIT"
},
{
"id": 6,
"parent": 3,
"text": "Our findings show advantage for the extraction procedure ,",
"relation": "FINDINGS"
},
{
"id": 7,
"parent": 6,
"text": "however a direct no - buffer approach might be an alternative ,",
"relation": "COMPARISON"
},
{
"id": 8,
"parent": 7,
"text": "since it identified up to 70 % of positive clinical specimens . <S> <P>",
"relation": "CAUSE-RESULT"
}
]
}
"<S>" and "<P>" denote the sentence and paragraph boundaries, respectively.
The blank entries (i.e., volume, pages, doi) in the bibtex item below will be filled in after publication in TACL.
@article{nishida2021outofdomain,
title={Out-of-Domain Discourse Dependency Parsing via Bootstrapping: {A}n Empirical Analysis on its Effectiveness and Limitation},
author={Nishida, Noriki and Matsumoto, Yuji},
journal={Transactions of the Association for Computational Linguistics},
volume={},
pages={},
year={2021},
doi={},
}