This package uses ideas from nonlinear dynamical system theory to model and interrogate agent state trajectories within games, biological associative memory, and switching autoregressive processes.
Authors: Nick Richardson and Yoni Maltsman
git clone https://github.com/njkrichardson/saddlecity.git
pip install -e saddlecity
Linear dynamical systems are simple but powerful abstractions for modelling time-evolving processes of interest. Despite their broad application domain, often the linear constraint on the evolution of a state vector lacks the capacity to appropriately model processes with sufficiently nonlinear structure. The domain of nonlinear dynamical systems provides a theoretical framework to reason about dynamical systems in which this linear constraint has been lifted.
This package showcases three applications of nonlinear dynamical system theory: simulations of agent state trajectories from game theory, computational models of biological memory systems, and switching autoregressive processes from the signal processing/stochastic processes literature.
The Hopfield network is a fully connected, unsupervised neural network designed to act as a model of associative memory. Here we summarize the distinguishing features between conventional digital memory and associative biological memory with a passage from David MacKay's Information Theory, Inference, and Learning Algorithms textbook.
- Biological memory is associative. Memory recall is content-addressable. Given a person’s name, we can often recall their face; and vice versa. Memories are apparently recalled spontaneously, not just at the request of some CPU.
- Biological memory recall is error-tolerant and robust.
- Errors in the cues for memory recall can be corrected. An example asks you to recall ‘An American politician who was very intelligent and whose politician father did not like broccoli’. Many people think of president Bush – even though one of the cues contains an error.
- Hardware faults can also be tolerated. Our brains are noisy lumps of meat that are in a continual state of change, with cells being damaged by natural processes, alcohol, and boxing. While the cells in our brains and the proteins in our cells are continually changing, many of our memories persist unaffected.
- Biological memory is parallel and distributed, not completely distributed throughout the whole brain: there does appear to be some functional specialization – but in the parts of the brain where memories are stored, it seems that many neurons participate in the storage of multiple mem- ories.
One can use saddlecity to instantiate general Hopfield networks.
from hopfield import Hopfield
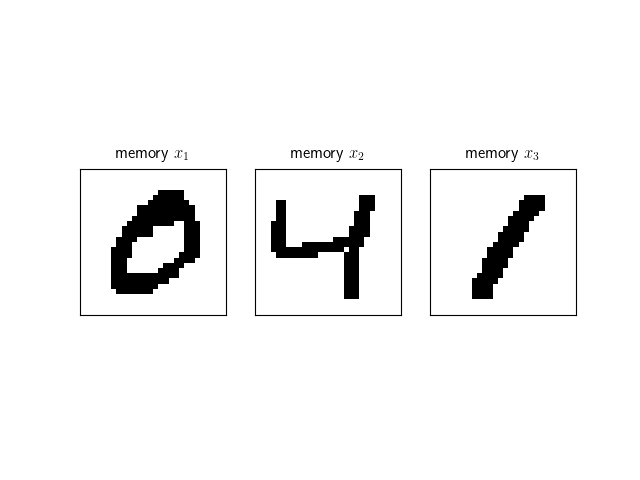
net = Hopfield() A network can be provided memories to memorize with the add_memories method. Here we add the following three memories corresponding to binary images of handwritten digits.
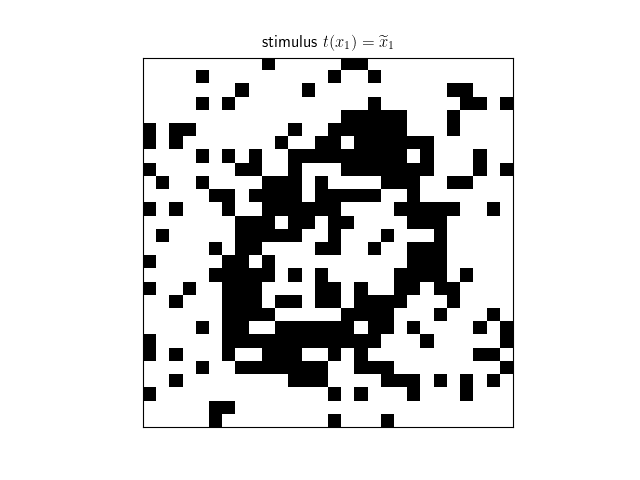
Recall that this is an unsupervised neural network model; and thus doesn't require any parameter estimation. We can now use the neural network to attempt to process and decode various stimuli corresponding to corrupted versions of the learned memories.
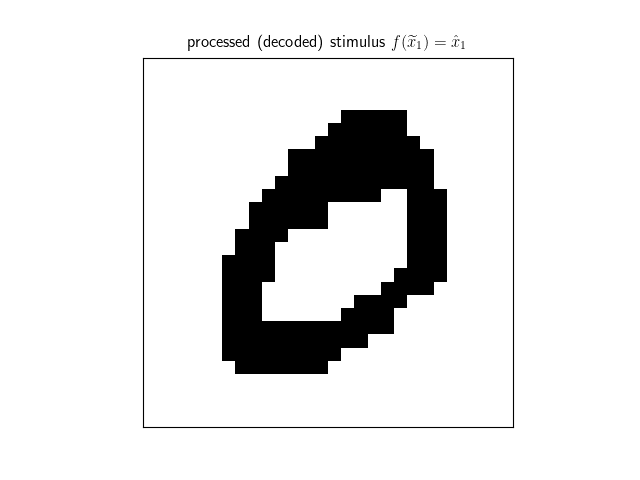
decoded = net.decode(stimulus) We can then visualize the processed stimulus, which has converged to the true memory corresponding to the stimulus.
This example and more can be found in the examples directory.
Text references and resources can be found under the resources directory. We follow David MacKay's text as a guiding reference on Hopfield networks, and David Barber's for a reference on switching autoregressive processes (both texts are freely available online).
We utilize Scott Linderman's Python package to implement state space models and demonstrate procedures for Bayesian learning of parameters in these models.