This repository contains the original implementation of "Align-gram : Rethinking the Skip-gram Model for Protein Sequence Analysis" in Tensorflow
Background: The inception of next generations sequencing technologies have exponentially increased the volume of biological sequence data. Protein sequences, being quoted as the ‘lan- guage of life’, has been analyzed for a multitude of applications and inferences.
Motivation: Owing to the rapid development of deep learning, in recent years there have been a number of breakthroughs in the domain of Natural Language Processing. Since these methods are capable of performing different tasks when trained with a sufficient amount of data, off-the-shelf models are used to perform various biological applications. In this study, we investigated the applicability of the popular Skip-gram model for protein sequence analysis and made an attempt to incorporate some biological insights into it.
Results: We propose a novel k-mer embedding scheme, Align-gram, which is capable of mapping the similar k-mers close to each other in a vector space. Furthermore, we experiment with other sequence-based protein representations and observe that the embeddings derived from Align-gram aids modeling and training deep learning models better. Our experiments with a simple baseline LSTM model and a much complex CNN model of DeepGoPlus shows the potential of Align-gram in performing different types of deep learning applications for protein sequence analysis.
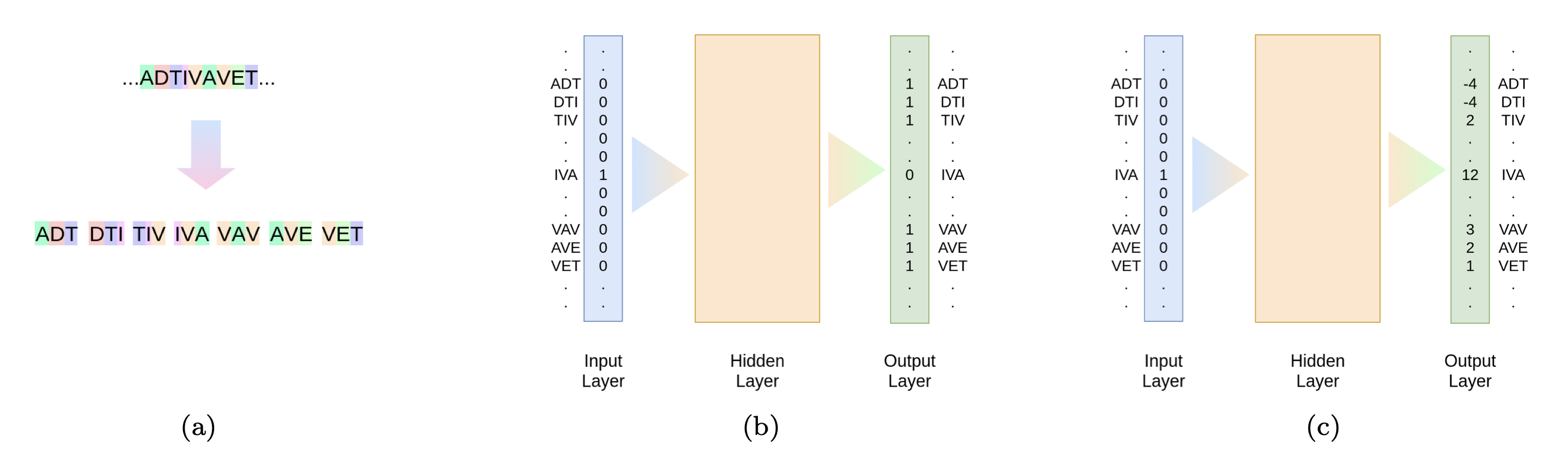
Fig. a presents breaking the sequence into 3-mer. In Skip-gram model (Fig. b) the 3-mers are then used to train the model using the proximity of the 3-mers. For the proposed Align-gram model (Fig. c) we compute the alignment scores between all the 3-mers and train the model to predict the alignment scores and in the process obtain the embedding matrix for 3-mers.
We provide the codes for Align-gram model and training. Our codes are based on tensorflow and numpy. The codes can be found in the /AlignGram directory.
- align_gram_model.py : contains the model definition
- model_builder.py : contains the codes for building, training and loading models.
- utils.py : contains the healper functions
You can train custom Align-Gram embedding based on your particular requirement.
All the steps involving model building and training is encapsulated in the buildModel function
Just input the value of k, embedding dimension, and select suitable substitution matrix and gap penalties based on your need (note: gap penalties need to be negative because of biopython). Also, provide a model_name which will be the filename for the saved model.
from AlignGram.model_builder import buildModel
model = buildModel(K, embedding_len, sub_mat, gap_open, gap_extend, model_name)
Next you can provide a protein sequence as input and compute it's embeddings
protein_seq = 'GIMHMEWLNISYIIHNQVFS'
protein_emb = model.computeProteinEmbedding(protein_seq)
You can also use the get_model function to load your model later.