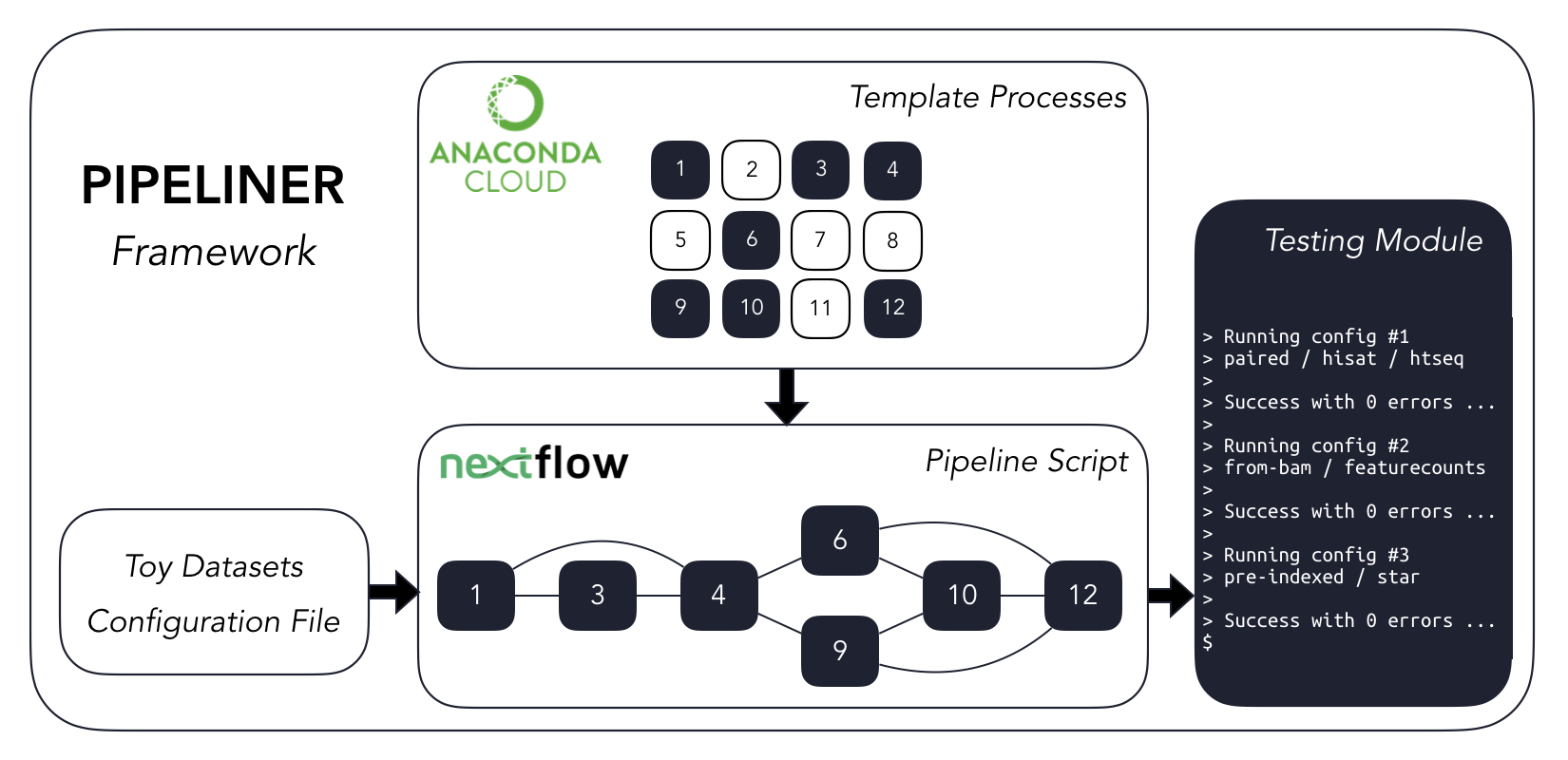
A flexible Nextflow-based framework for the definition of sequencing data processing pipelines
- Modular directory structure: It is designed to generate automated result directory based on the names of the samples and tools used to process them.
- Platform independent: It is bundled with an anaconda repository which contains pre-compiled tools as well as pre-built environments that can be used directly.
- Modular architecture: It allows the expert users to customize, modify processes, or add additional tools based on their needs.
- Automated job parallelization, job recovery, and reproducibility.
For more information, please refer to the full documentation
$ git clone https://github.com/montilab/pipeliner
$ conda env create -f pipeliner/envs/linux_env.yml # Linux$ conda env create -f pipeliner/envs/osx_env.yml # Mac$ source activate pipeliner$ python pipeliner/scripts/paths.py$ cd pipeliner/pipelines
$ curl -s https://get.nextflow.io | bash
$ ./nextflow rnaseq.nf -c rnaseq.configN E X T F L O W ~ version 0.31.1
Launching `rnaseq.nf` [nasty_pauling] - revision: cd3f572ab2
[warm up] executor > local
[31/1b2066] Submitted process > pre_fastqc (ggal_alpha)
[23/de6d60] Submitted process > pre_fastqc (ggal_theta)
[7c/28ee53] Submitted process > pre_fastqc (ggal_gamma)
[97/9ad6c1] Submitted process > check_reads (ggal_alpha)
[ab/c3eedf] Submitted process > check_reads (ggal_theta)
[2d/050633] Submitted process > check_reads (ggal_gamma)
[1d/f3af6d] Submitted process > pre_multiqc
[32/b1db1d] Submitted process > hisat_indexing (genome_reference.fa)
[3b/d93c6d] Submitted process > trim_galore (ggal_alpha)
[9c/3fa50b] Submitted process > trim_galore (ggal_theta)
[62/25fce0] Submitted process > trim_galore (ggal_gamma)
[66/ccc9db] Submitted process > hisat_mapping (ggal_alpha)
[28/69fff5] Submitted process > hisat_mapping (ggal_theta)
[5c/5ed2b6] Submitted process > hisat_mapping (ggal_gamma)
[b4/e559ab] Submitted process > gtftobed (genome_annotation.gtf)
[bc/6f490c] Submitted process > rseqc (ggal_alpha)
[71/80aa9e] Submitted process > rseqc (ggal_theta)
[17/ca0d9f] Submitted process > rseqc (ggal_gamma)
[d7/7d391b] Submitted process > counting (ggal_alpha)
[df/936854] Submitted process > counting (ggal_theta)
[11/143c2c] Submitted process > counting (ggal_gamma)
[31/4c11f9] Submitted process > expression_matrix
[1f/3af548] Submitted process > multiqc
Success: Pipeline Completed!
Please refer to reports for examples
Federico A, Karagiannis T, Karri K, Kishore D, Koga Y, Campbell J, Monti S (2019) Pipeliner: A Nextflow-based framework for the definition of sequencing data processing pipelines. Frontiers in Genetics. https://doi.org/10.3389/fgene.2019.00614.