Top-5% solution to the BMS Molecular Translation Kaggle competition on chemical image-to-text translation.
Organic chemists frequently draw molecular work using structural graph notations. As a result, decades of scanned publications and medical documents contain drawings not annotated with chemical formulas. Time-consuming manual work of experts is required to reliably convert such images into machine-readable formulas. Automated recognition of optical chemical structures with deep learning helps to speed up research and development in the field.
The goal of this project is to develop a deep learning framework for chemical image captioning. In other words, the project aims at translating unlabeled chemical images into the text formulas. To do that, I work with a large dataset of more than 4 million chemical images provided by Bristol-Myers Squibb.
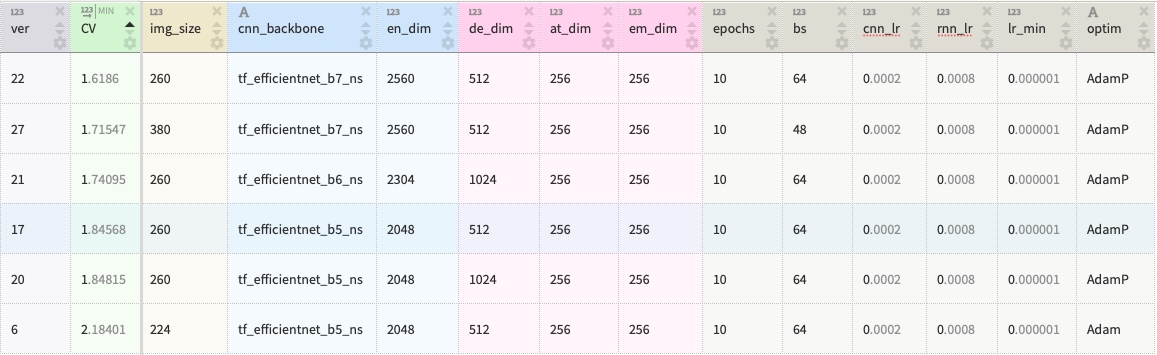
My solution is an ensemble of seven CNN-LSTM Encoder-Decoder models implemented in PyTorch. The table below summarizes the architectures and main training parameters. The solution reaches the test score of 1.31 Levenstein Distance and places in the top-5% of the competition leaderboard. The detailed summary is provided in this writeup.
The project has the following structure:
codes/:.pymain codes with data, model, training and inference modulesnotebooks/:.ipynbColab-friendly notebooks for data augmentation and model traininginput/: input data (not included due to size constraints, can be downloaded here)output/: model weights, configurations and figures exported from the notebooks
To work with the repo, I recommend to create a virtual Conda environment from the environment.yml file:
conda env create --name bms --file environment.yml
conda activate bms
The solution can then be reproduced in the following steps:
- Download competition data and place it in the
input/folder. - Run
01_preprocessing_v1.ipynbto preprocess the data and define a chemical tokenizer. - Run
02_gen_extra_data.ipynband03_preprocessing_v2.ipynbto construct additional synthetic images. - Run training notebooks
04_model_v6.ipynb-10_model_v33.ipynbto obtain weights of base models. - Perform normalization of model predictions using
11_normalization.ipynb. - Run the ensembling notebook
12_ensembling.ipynbto obtain the final predictions.
All training notebooks have the same structure and differ in model/data parameters. Different versions are included to ensure reproducibility. To understand the training process, it is sufficient to go through the codes/ and inspect one of the modeling notebooks. The ensembling code is also provided in this Kaggle notebook.
More details are provided in the documentation within the codes & notebooks.