Labeling C. elegans Neurons from NeuroPAL Images
GUI environment
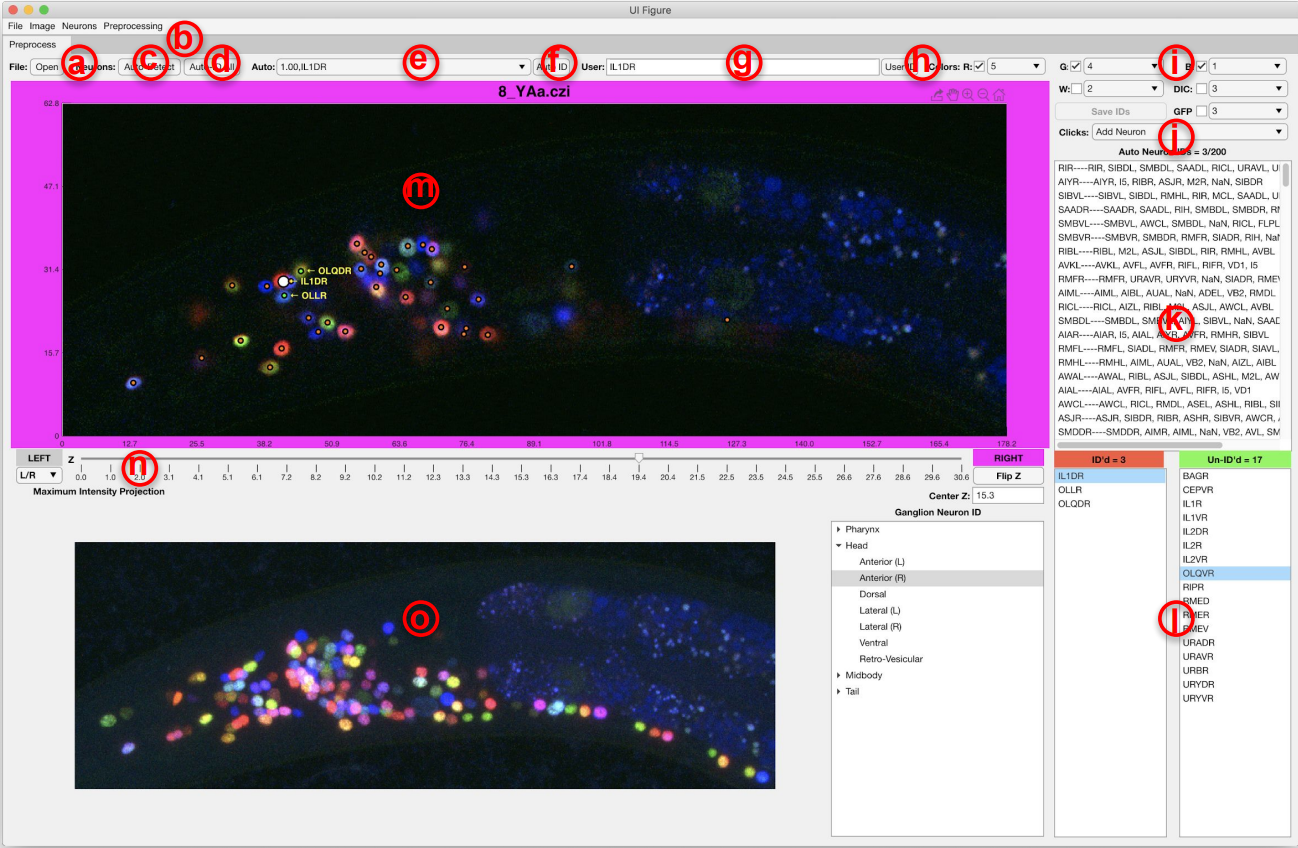
(a) open and load a NeuroPAL image, (b) menu for basic preprocessing and improving the visualization, (c) automatically detect the neurons in NeuroPAL images, (d) automatically identify and label the detected neurons (e) clicking on each neurons shows the potential names that our method has found for that neuron and their associated uncertainties, (f) confirm the neuron identity, (g,h) manually annotate neurons, (i) selecting color channels used for visualizing NeuroPAL image, (j) select double click action: manually add a neuron/automatically find a neuron near the clicked location/remove artefacts, (k) list of neuron names sorted by their uncertainty according to the result of our method, (l) true neuron names and colors categorized by their corresponding ganglia, (m) z-stacks of NeuroPAL image and text labels for annotated or confirmed neurons, (n) controller for moving through different z-stacks, (o) maximum intensity projection of the image.
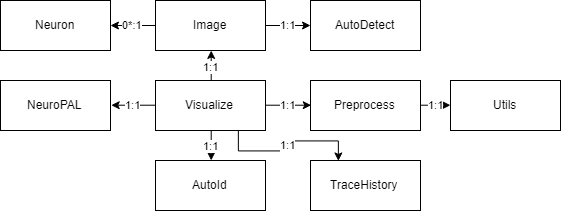
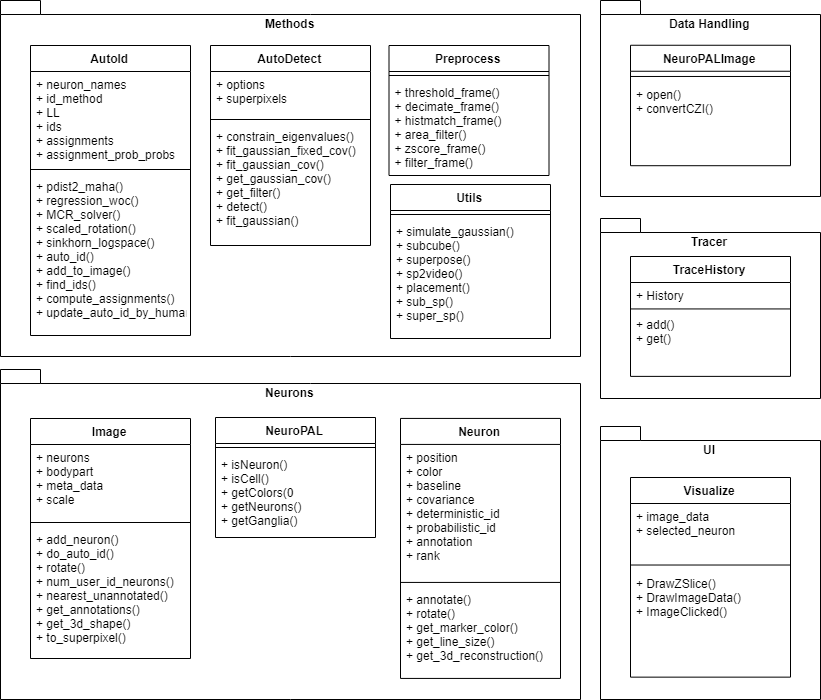
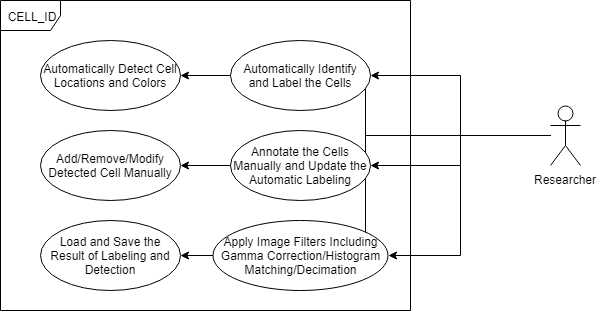
Class, Package, and Usecase Diagrams
To build a fast and scalable system we followed object oriented design practices. We identified internally coherent and externally independent classes in our environment and established their properties and behavior, as well as their interactions with other modules. We have separate packages for data handling, methods, logging, biological objects, and user interface. Each of these packages contains relevant classes and the functionalities of the system is built upon the interactions between the classes. For reusability purposes we tried to develop a modular system with independent modules. This allows the users of the system to reuse different compartments of the system for other purposes.
Related article
For more details about the experimental setup and the methods use the NeuroPAL paper:
@article{PMID:33378642, Title= {NeuroPAL: A Multicolor Atlas for Whole-Brain Neuronal Identification in C. elegans}, Author= {Yemini, Eviatar and Lin, Albert and Nejatbakhsh, Amin and Varol, Erdem and Sun, Ruoxi and Mena, Gonzalo E and Samuel, Aravinthan D T and Paninski, Liam and Venkatachalam, Vivek and Hobert, Oliver}, DOI= {10.1016/j.cell.2020.12.012}, Number= {1}, Volume= {184}, Month= {January}, Year= {2021}, Journal= {Cell}, ISSN= {0092-8674}, Pages= {272—288.e11}, Abstract= {Comprehensively resolving neuronal identities in whole-brain images is a major challenge. We achieve this in C. elegans by engineering a multicolor transgene called NeuroPAL (a neuronal polychromatic atlas of landmarks). NeuroPAL worms share a stereotypical multicolor fluorescence map for the entire hermaphrodite nervous system that resolves all neuronal identities. Neurons labeled with NeuroPAL do not exhibit fluorescence in the green, cyan, or yellow emission channels, allowing the transgene to be used with numerous reporters of gene expression or neuronal dynamics. We showcase three applications that leverage NeuroPAL for nervous-system-wide neuronal identification. First, we determine the brainwide expression patterns of all metabotropic receptors for acetylcholine, GABA, and glutamate, completing a map of this communication network. Second, we uncover changes in cell fate caused by transcription factor mutations. Third, we record brainwide activity in response to attractive and repulsive chemosensory cues, characterizing multimodal coding for these stimuli.}, URL= {https://doi.org/10.1016/j.cell.2020.12.012}}
Here is a link to publication: https://www.cell.com/cell/fulltext/S0092-8674(20)31682-2
To open NeuroPAL images the following packages needs to be installed:
https://docs.openmicroscopy.org/bio-formats/5.3.4/users/matlab/index.html https://github.com/StackOverflowMATLABchat/mlapptools
We also use the following MATLAB functions but we have included them in the software:
https://github.com/tminka/lightspeed/blob/master/logsumexp.m https://www.mathworks.com/matlabcentral/fileexchange/20328-munkres-assignment-algorithm