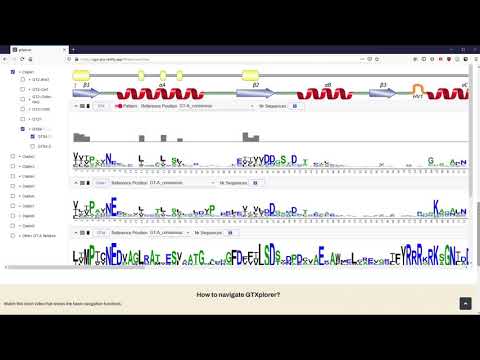
GTXplorer: A portal to navigate and visualize the evolutionary information encoded in fold A glycosyltransferases
This repo includes the GTXplorer web database files and can be downloaded to deploy a local copy of GTXplorer.
To change labels, and visibility of the controls, you may edit the web-react/src/gta.settings.json file.
-
by clicking on the green
codebutton at the top to download as zip or use the following command:
git clone https://github.com/esbgkannan/GTxplorer.git -
cd web-reactnpm install(if necessary, runrm -rf node_modulesbefore this command)npm start(it starts the server on http://localhost:3000, you can change the port in theweb-react/src/package.jsonfile).
-
-
cd web(Optional): If the local server is running on an address except http://localhost:3000,
-
Edit the
srcproperty value in the following line inindex.html:<iframe id="react_frame" src="http://localhost:3000/" width="550" height="550" frameborder="0" scrolling="yes" style="width: 100%"> -
Edit the
SERVER_ADDRESSvariable value inweb/src/JS/index.js
-
-
Start the project on a local server:
serve -s .Note: If
serveis not installed. Install it by runningnpm install -g serve.Now, you can navigate to
http://localhost:5000/to view the website.
-
-
Get the data files that are available for download through the web version of GTXplorer using the following script. Run the script from the GTXplorer folder.
cd GTxplorer
bash download.sh
All the complete datasets for GTXplorer are in the db_files folder.
-
Fasta/
Includes individual files per GTA clade, family and subfamily with all the GTA domain sequences for that level.
-
FastaFull/
Includes individual files per GTA clade, family and subfamily with all the GTA full length sequences for that level.
-
Tables/
Contains tab delimited files with extensive information for all GTA sequences in a given level (clade,family or subfamily).
-
Domains/
For each of the 53 GTA families, this directory contains tables with the domain organization information (one shortened easily readable version and an extended version with more detailed information).
-
Tax/
This directory includes files with taxonomic distribution information for all GTA clades, families and subfamilies.
-
WeblogoDomain/
Includes png images of the weblogs for the alignments of sequences from GTA clades, families and subfamilies.
-
Hierarchy.tsv
Tab delimited file defining the entire hierarchy of the GTA fold.
-
GTA_Tree.csv
A csv file for the generation of KinView style viewer.
-
GTA_Clade.json
Includes details to display in the cards for the 9 major clades.
-
GTA_Fam.json
Includes details to display for the 53 major GTA families (in the labels of the GTA tree)
-
GTA_SubFam.json
Incudes details for the GTA subfamilies.
Natarajan Kannan: nkannan@uga.edu
Rahil Taujale: rtaujale@uga.edu
- v1.0.0
- First packaged beta version.
If you find this tool helpful, please cite:
Rahil Taujale, Saber Soleymani, Amitabh Priyadarshi, Aarya Venkat, Wayland Yeung, Krys Kochut and Natarajan Kannan. GTXplorer: A portal to navigate and visualize the evolutionary information encoded in fold A glycosyltransferases. Preprint.
This work is licensed under the MIT license.