A R package to analyze data from different massively parallel reporter assays
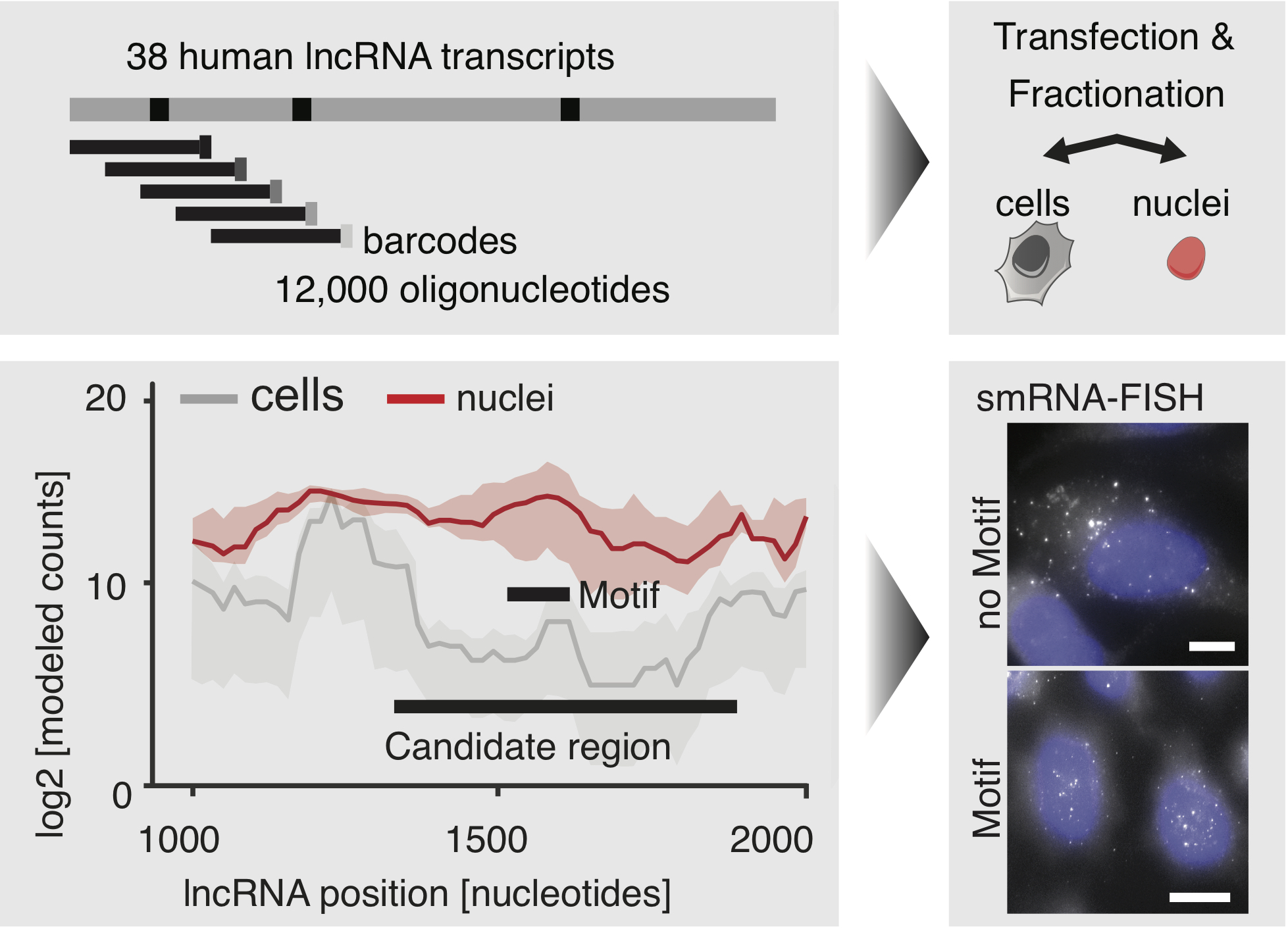
Since the early days of molecular biology, scientists have used reporter assays for different applications including studying the regulation of gene expression. While conventionally, one regulatory element is assayed at a given time, recent breakthroughs in microarray based nucleotide synthesis and next generation sequencing have allowed scientists to test several thousand elements in a single massively parallel reporter assay (MPRA). MPRA's were first demonstrated in Patwardhan R.P. et al. (2009) and Melnikov A. et al. (2012) and the underlying technology has only become stronger and more robust in the following years. Recently, there has been a flurry of papers which have used MPRA's to test potential enhancer elements, eQTL's, SNP's as well as repressive elements - Ulirsch J.C. et al. (2016), Tewhey R. et al (2016), Ernst J. et al. (2016). Although many "games" are being played by clever scientists to deduce regulatory elements using MPRAs, currently there is no dedicated software for analyzing MPRAs - which often have constraints a bit different from other *-Seq technologies. This is especially true for games which tile a long region of interest with short oligos and aim to identify regulatory elements contained within. Here, we provide the user functions for all aspects of MPRA experiments - from design of the oligo pool to learning kmers enriched in the infered regulatory elements.
The following functions are provided in the package:
- designOligoPool - Input a FASTA file of the regions you want to tile and get an output oligo arrary
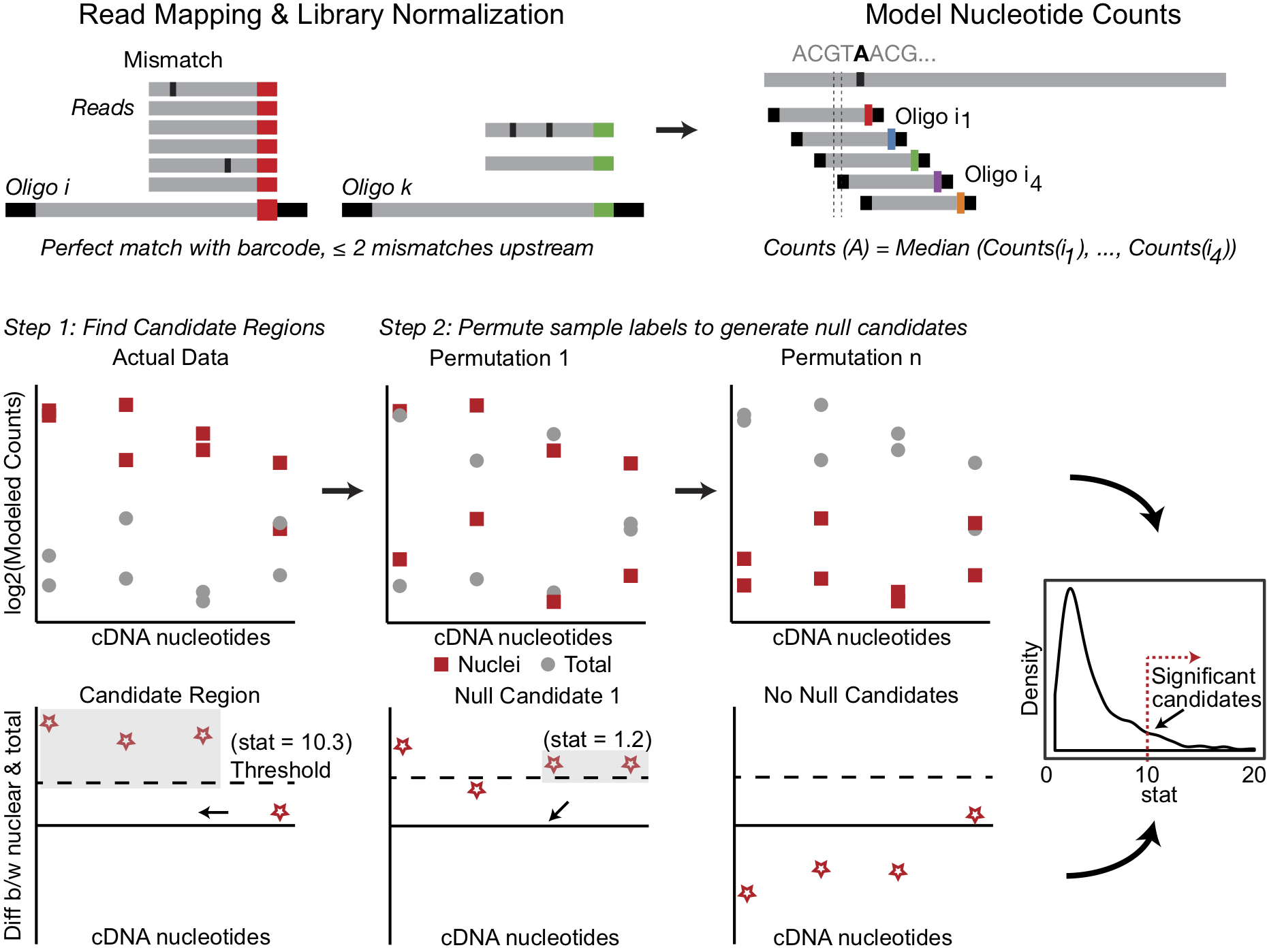
- mapToBarcodes - Map the FASTQ files from your experiment to the barcodes and generate a table of counts for each oligo
- normCounts - Normalize oligo counts based on library size
- modelNucCounts - Model nucleotide level counts from normalized oligo level counts
- DRfinder - Perform inference to detect differential regions in oligo experiments
Please browse the vigenttes or read the documentation to understand the learn all the available options.
Currently, oligoGames is under development and we recommend you contact us here before using it.
The easiest way to install the development version directly from Github is
devtools::install_github("cshukla/oligoGames")After installation, you can load the package into R.
library(oligoGames)Now you are ready to play some games :)
Here, we describe how to play a demo game. To begin playing, we first need to design an oligo pool.
We do this by using the designOligoPool function. We need a fasta file of sequences which are interested in tiling. Once we have that file, it is very simple to generate our oligo pool.
regionsFile <- system.file('extdata', 'testRegions.fa' package='oligoGames')
microSeedsFile <- system.file('extdata', 'hg19miRSeeds.txt.gz', package='oligoGames')
outDir <- 'demoOligoGame'
designOligoPool(regionsFile, microSeedsFile, outDir)At this stage, we need to order the oligos and do the experiment (likely several times because it never works the first time). Since this is a demo game, we are going to fast forward the 2.5 long years which we spent doing this and assume we have the sequencing data.
Next, we map the data to our oligo barcodes with the mapToBarcodes function.
fastqCases <- c(system.file('extdata', 'fastqFiles', 'case1.fastq.gz', package='oligoGames'),
system.file('extdata', 'fastqFiles', 'case2.fastq.gz', package='oligoGames'))
fastqControl <- c(system.file('extdata', 'fastqFiles', 'control1.fastq.gz', package='oligoGames'),
system.file('extdata', 'fastqFiles', 'control2.fastq.gz', package='oligoGames'))
oligoMap <- system.file('extdata', 'lncLocOligoPool.fa' package='oligoGames')
mapToBarcodes(fastqCases, fastqControls, oligoMap, demoOligoGame)
Our next step is to normalize the counts for library size. We use the normCounts function for this. Please note that we only need to provide the location of the unnormalized counts. The function automatically reads this file and returns a normalized counts matrix.
rawCounts <- system.file("extdata", "allTranscriptsCounts_Raw.tsv", package = "oligoGames")
rawCounts <- utils::read.table(rawCounts, header=TRUE, stringsAsFactors=FALSE)
normalizedCounts <- normalize(rawCounts, normType='median')We will now go from oligo counts to nucleotide level counts using modelNucCounts. One thing to note here is that the name of the transcripts should be similarly formated as thed default data shown below. Read the documentation of modelNucCounts for more info.
Also, similar to the unnormalized counts file you only need to provide a path to your meta data file. The function will automatically open and read the file.
metaData <- system.file("extdata", "oligoMeta.tsv", package = "oligoGames")
conditionLabels <- c("Nuclei", "Total")
oligoLen <- 110
modeledNucs <- modelNucCounts(normalizedCounts, metaData, conditionLabels, modelMethod = "median", oligoLen)Now we are in the final round of our game and we will infer the differential regions with the help of DRfinder
DRregions <- DRfinder(modeledNucs, conditionLabels, minInSpan = 5,
bpSpan = 50, minNumRegion = 3, cutoff = 0.05, smooth = TRUE,
verbose = TRUE, workers = 1, maxPerms = 50)Report bugs as issues on the GitHub repository