Source code for malware can easily be had by googling, searching on pastebin or running a WordPress honey pot. Much of it is interpreted languages, like PHP or Perl.
I will use "Web Shell by oRb" (a.k.a "WSO", "FilesMan") and the omnipresent IRC Perl bot (Perl/Shellbot) as examples.
"Web Shell by oRb" (also called "WSO", "FilesMan" and "AntiChat shell") is by far the most popular web shell downloaded to hacked CMS web sites today. It's often reformatted, obfuscated or otherwise slightly customized before hiding it in a WordPress installation. Many distinct versions exist, including one under active development.
The omnipresent IRC Perl bot is referred to by antivirus firms as "Perl/Shellbot" or some form of that indicating a derivative. It's a piece of Perl code that signs in to specific IRC servers and channels, and listens for commands from its botherder. A typical variant can be had from Pastebin.
Casual reading of several variants of either WSO or Perl/Shellbot will convince you that the variants are "related". It seems that one variant "evolved" into another via a process of spammers making small changes to suit their needs. Perhaps a selection of the fittest happens when one script kiddy copys the succssful code of another script kiddy. Collection and comparison of variants should allow the construction of a "family tree" of variants. This project is a description and demonstration of one method of constructing the family tree, or phylogeny, of WSO, and less succesfully, of Perl/Shellbot.
Ordinary biological evolution analogies are ill-suited to reconstructing malware phylogenies. Biological phylogenies assume binary splits between two clades, and assume that we know the sister clades, but can't identify the the common ancestor. Malware can split many times from a single, identifiable common ancester. Biological phylogenies assume a single common ancestor. Malware often, maybe almost always, has more than a single ancestor.
Souce code malware seems to evolve by feature addition or subtraction. People modifiying malware add small chunks of code, code that might phone home with URL, user ID and password, or just evaluate some code when executed with a special CGI-BIN parameter. Sometimes entire functions or subsystems are transplanted from another program.
When the "source code" (DNA) of biological families is available, it rarely shows evolution by adding or subtracting entire "features". Biological evolution appears to happen by gradual changes over large extents of a clade's DNA. The closest biological analogy of the way source code malware evolves would seem to be horizontal gene transfer.
Traditional biology phylogenies are sometimes calculated with a "distance" metric, and tree construction algorithms like Neighbor Joining. This section illustrates such a calculation.
I used the NCD algorithm from Common Pitfalls Using The Normalized Compression Distance
it seems to give closer-to-zero "distance" of a piece of code to itself. The code is in
distance_calc.php
NCD(x,y) = max(C(xy) - C(x), C(yx) - C(y)}/max(C(x), C(y))
NCD(x,y) is the normalized compression distance from string x to string y,
max(a, b) is the numerical maximum of numbers a and b, and C(e) is the
size in bytes of compressed string e. xy is string x with string y
appended. I used PHP's bzcompress() function. File distance_calc.php
implements the NCD calculations.
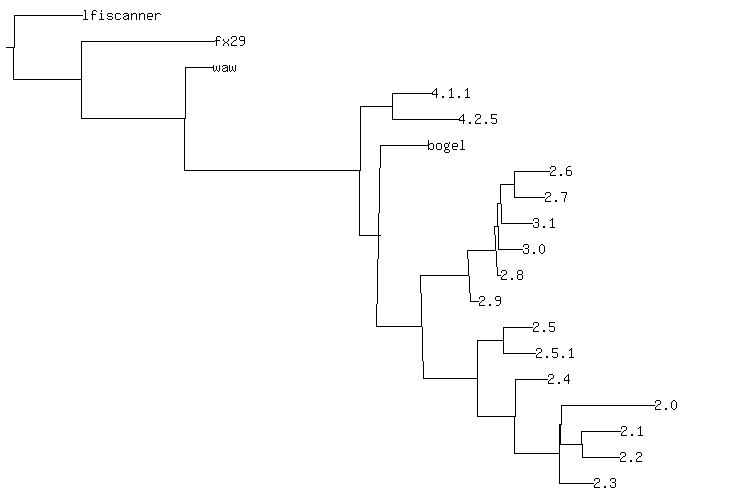
The following phylogeny is for a group of versions of "Web Shell by oRb" (WSO), described
futher below, except for lfiscanner.php and fx29, which are secondary ancestors
of some versions of WSO.
All WSO versions are files in exemplars/, PHP source code, normalized by pretty
printing, and removing ASCII carriage
return characters. I did an NCD calculation on all files in exemplars.
Using the neighbor joining algorithm, I calculated the following phylogeny.
You pick an "outgroup" to get a rooted tree from neighbor joining. I
chose lfiscanner.php.
Horizontal lengths of branches are proportional to the normalized
compression distance separating source files. You can re-generate the
above image by invoking make nj_phylogeny.png. The file makefile
describes how to rebuild nj_phylogeny.png from scratch.
NCD and neighbor joining get some parts of the phylogeny correct. Although I
chose lfiscanner as the out group, it correctly found the fx29 web shell as
the next most distantly-related piece of code. It gets the basic split between
WSO 2.4 and WSO 2.5 clades, but mixes version 2.8 and 3.1 in the 2.4-based
clade - version 2.8 is clearly a minor hack of version 2.5.1, differing only by
yet another phone home. Version 3.1 descends from 2.5 by the verbatim
incorporation of a local-file-include scanner from lfiscanner.php.
The assumptions inherent in neighbor joining make version 2.4 into an outgroup
of a clade. In addition to comprising the immediate ancestor of a number of
species, version 2.4 should appear as the ancestor of a linear progression of
2.5 to 2.5.1 to 2.8. The entire issue of multiple ancestry doesn't appear at
all. fx29 and lfiscanner.php should show up as auxilliary ancestors to
multiple variants that have a WSO variant as a primary ancestor.
All told, this phylogeny is roughly correct, but has too many flaws to be more than a general guide.
Because malware evolves by feature addition and subtraction, and not by changes in genomes, I tried to use the pieces of source code that differ between species, and the pieces of source code that remain common between all members of a clade to derive a phylogeny. I explictly allow for multiple ancestry.
We need to distinguish between feature and trait for the following. I use "feature"
to mean some distinguishable action the source code carries out, for example storing
application state in an HTTP cookie. I use "trait" for the code fragment chosen
to represent this feature: function WSOsetcookie.
Determining which traits to use is important, and not amenable to automation. The algorithm needs at least one trait (line or lines of source code) common between an ancestor and all of its desendants. It will calculate multiple independent trees if this condition is not met.
The algorithm needs each pair of species to differ in at least one feature, and descendants must differ from immediate ancestors by at least one feature.
Species are represented by instances of a data structure that can contain a set
of traits, and allow each trait to be marked. Sets of traits in
two species must be comparable to determine a subset relationship. The data
structure must be able to keep references to immediate descendant species. The
data structure must be capable of being marked to indicate that it has at least
one ancestor. This project has such a data structure in file phylogeny/Species.php
- Start with an empty set of root species, and a separate empty list of species.
- Order species in a list by decreasing number of traits.
- Remove species with largest number of traits from ordered species list. This species becomes the current species.
- Compare current species featuries to next species' traits in decreasing order of number of traits.
- When a species on the list has traits that are a strict subset of current species' traits list, and contain a trait previously unmarked in the current species, assign current species as child of other species.
- Mark all traits of the current species that are common to the new parent.
- If more than one trait is unmarked, repeat from (3)
- If no more than one trait is unmarked, go to (2).
- If you have compared traits of the current species to all species left on the list, and the current species hasn't been added to any species as a child, add the current species to the set of root species. Go to (2)
- When the ordered list is empty, stop.
- Starting from the set of root species created in step (8), use each root node's children to draw the resulting phylogeny.
I implemented the algorithm in file phylogeny/p2.php in this repository.
The algorithm above always terminates. Every species gets popped off ordered list of species, and there's only a finite number of species on that ordered list. There's only a finite number of comparisons to other species made for each current species.
Consider a phylogenetic tree with one root, many children, and no
grandchildren. Only one trait differs between any two child species.
The first of the N species gets compared to N - 1 child species before being assigned
to the root species. The second species gets compared to N - 2 species, the third
species gets compared to N - 3 species, and so forth. The number of species-to-species
comparisons is:
(N-1)+(N-2)+(N-3)+...+(N - (N-2))(N - (N-1))
This is just N things taken two at a time, which is: N!/(2!(N - 2)!) This formula reduces to N(N-1)/2 or (N2 - N)/2. Thus the algorithm takes O(N2) in the worst case.
Two species without at least one trait to differentiate them show up in a line of descent. The order is dependent on the sort algorithm used in step (1) above. Correctly chosing traits could cause them to show up as sister species.
Using a trait that's not in the real ancester for one of two sister species can lead to inversions of descent.
Mistaking a primitive trait as a trait added later can cause false sister clades, when in reality, one species of one of the clades is the ancestor of the other clade.
WSO is a "web shell", a single PHP file, invoked by a web browser, that can perform system administration tasks. It can examine system configuration, manipulate files, upload or download files. It also has "hacker" oriented features, like MySQL and Postgres database manipulation, remote code execution, password guessing and string format manipulation.
One of the interesting things about WSO code is that the original author chose to include a version string, "2.0" or "2.5" or "3.1". This gives a natural trait to do initial categorization of files into versions. The code versions 2.0 through 2.5 looks like a diligent developer wrote it. Each version has bugs fixed and features added over the version before. The versions other than 2.0 through 2.5 are quite different. They don't include bug fixes, but rather include whole chunks of code. These chunks range from quietly emailing home whenever that version is run, to "mass defacing" or "local file include" functions. No bugs get fixed, but the version numbering miraculously stays independent, in that there aren't two clades of WSO both version numbered 2.8, for example. Evolution by feature addition is obviously the mode of development outside of the version 2.0 through 2.5 progression. Version 2.5 is the only code I have confidence of possessing the original.
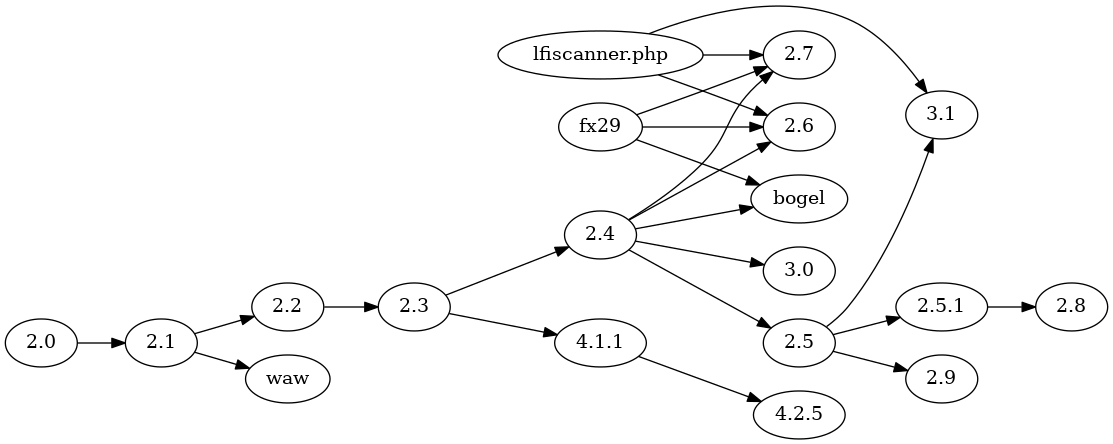
Above is a phylogeny for WSO versions using the algorithm in this project.
You should be able to reproduce the phylogeny above by cloning this repository,
and issuing a make wso_phylogeny.png command.
Compare this phylogeny to the one calculated with NCD and neighbor joining above. Version 2.4
constitutes the ancestor of many versions. The waw version is revealed as
a modification of version 2.1, rather than as a sister clade of all other versions.
My selection and enumeration of features of WSO is in the script find_traits.
I determined the features by careful manual code examination. If you read
find_traits and the WSO exemplars carefully, you will notice that I've done
some clever trait selection. Every version and variant up to 2.4 (and 2.4
descendants not related to 2.5) keeps information in a PHP session variable,
which resides on the server. Versions 2.5 and descendants (2.5.1, 2.8, 3.1) keep the same
information in HTTP cookies, presumably to avoid leaving traces behind on the
exploited server. I did not use "use PHP session" as a feature, but I did
use "use HTTP cookies" as a feature. This works around the algorithm not
accounting for feature deletions, just additions. Workarounds may not be
possible in all such situations.
Why does WSO appear in so many versions? "oRb" wrote the code in a mostly modular fashion.
"oRb" actually designed the program. This makes it easier to add backdoors or phone-homes,
change password, modify page headers and footers, or add new functions.
This is in contrast to other web shells like fx29, c99 or r57. These other web shells
are an unholy mess inside, with little or no modularity, and no easy way to add functionality.
"oRb" also shepherded WSO through a corporate-style linear improvement process from versions 2.0
through 2.5
Perl/Shellbot is a piece of Perl that understands the IRC protocol. Specially formatted messages from the botherder cause the bot to do things, like perform a UDP packet flood, run a command via the system's shell, or look up "dorks" in every search engine on the planet. The code arrives in a dazzling number of variants, some with function names and commentary in Portugese, others with function names & etc in English. Every botherder seems to do minor customization.
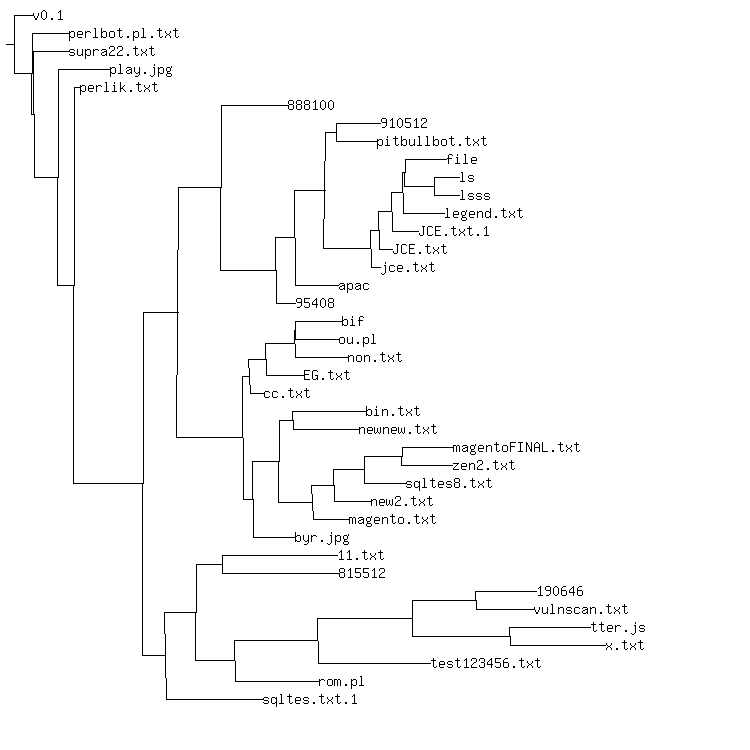
Above, a biology-inspired phylogeny using NCD and neighbor joining for 39
Perl/Shellbot variants I collected, or had downloaded to a honey pot. I did select
the v0.1 variant as the outgroup to construct this tree. Neighbor joining did catch
perlbot.pl.txt as the next most ancestral version, which seems correct, as perlbot.pl.txt
seems almost identical, barring some extra comments.
The really wrong thing that NCD/NJ does is mix the English variants in with the
Portugese variants. English versions new.txt.2, STMIK2.txt and
sqltes.txt.1 all get placed near the base of the phylogeny. Careful examination
of the Shellbot variants convinced me that the Portugese commented and named
variants are primitive, while the English variants are derivative. Moreover, the
English variants are quite a bit more uniform than the Portugese variants, all
containing some variant of the search engine "dork" lookup code. All English
variants also lack the package DCC code that's in some of the Portugese
variants, including the most primitive.
I combed through all 39 of the Perl/Shellbot variants for features and traits.
The simplest variants seem to have Portugese function names and variables names,
but at the same time, seem to be patched together from other programs and modules.
For instance, sub fixaddr has a Portugese comment that has it coming from
Perl module Net::IRC::DCC.
The variants with English language comments and function names seem far more uniform.
Every version (except byr.jpg) includes code for searching Google and other
search engines for text fragments. The Portugese variants exhibit a lot more peculiarities.
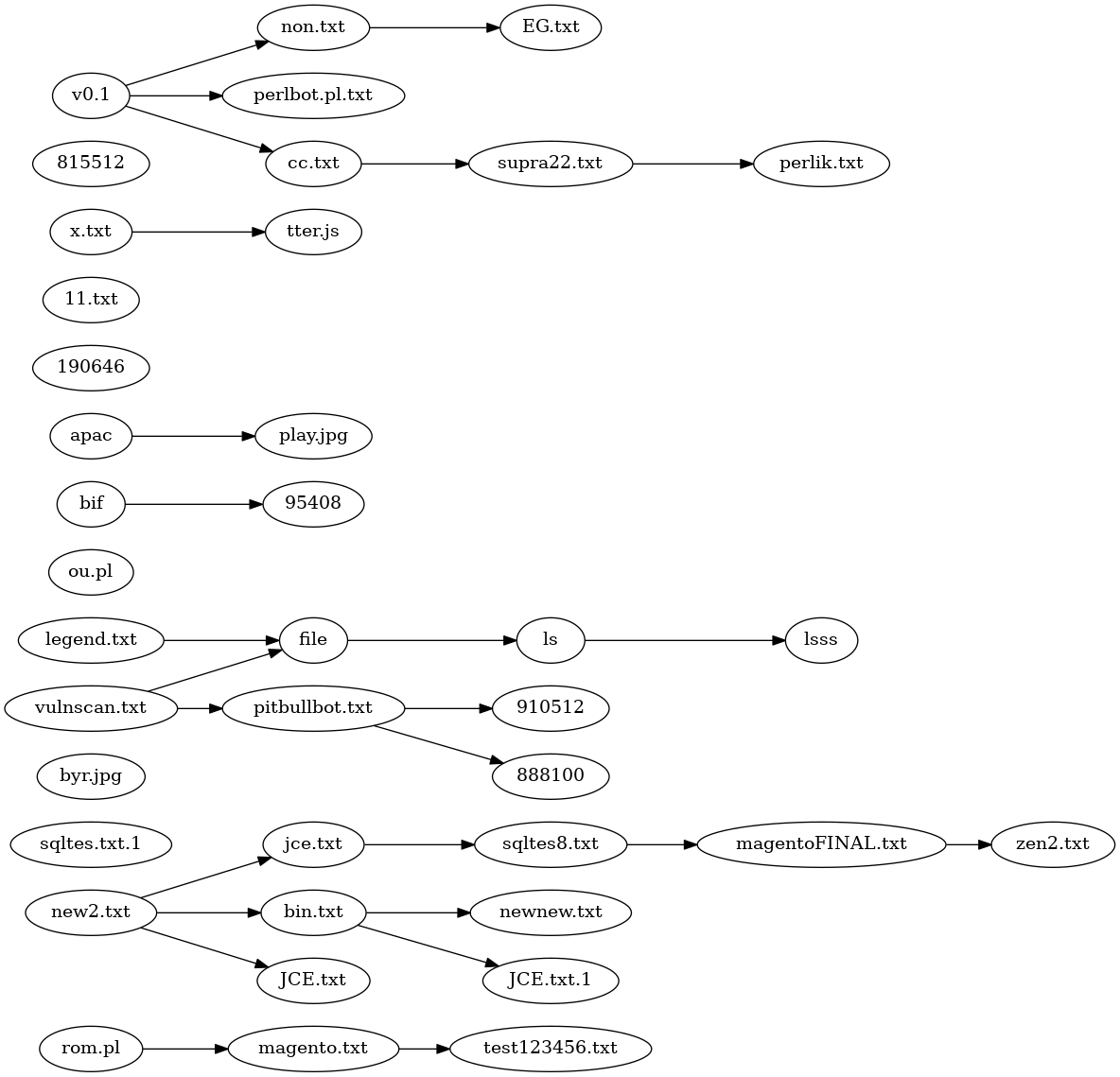
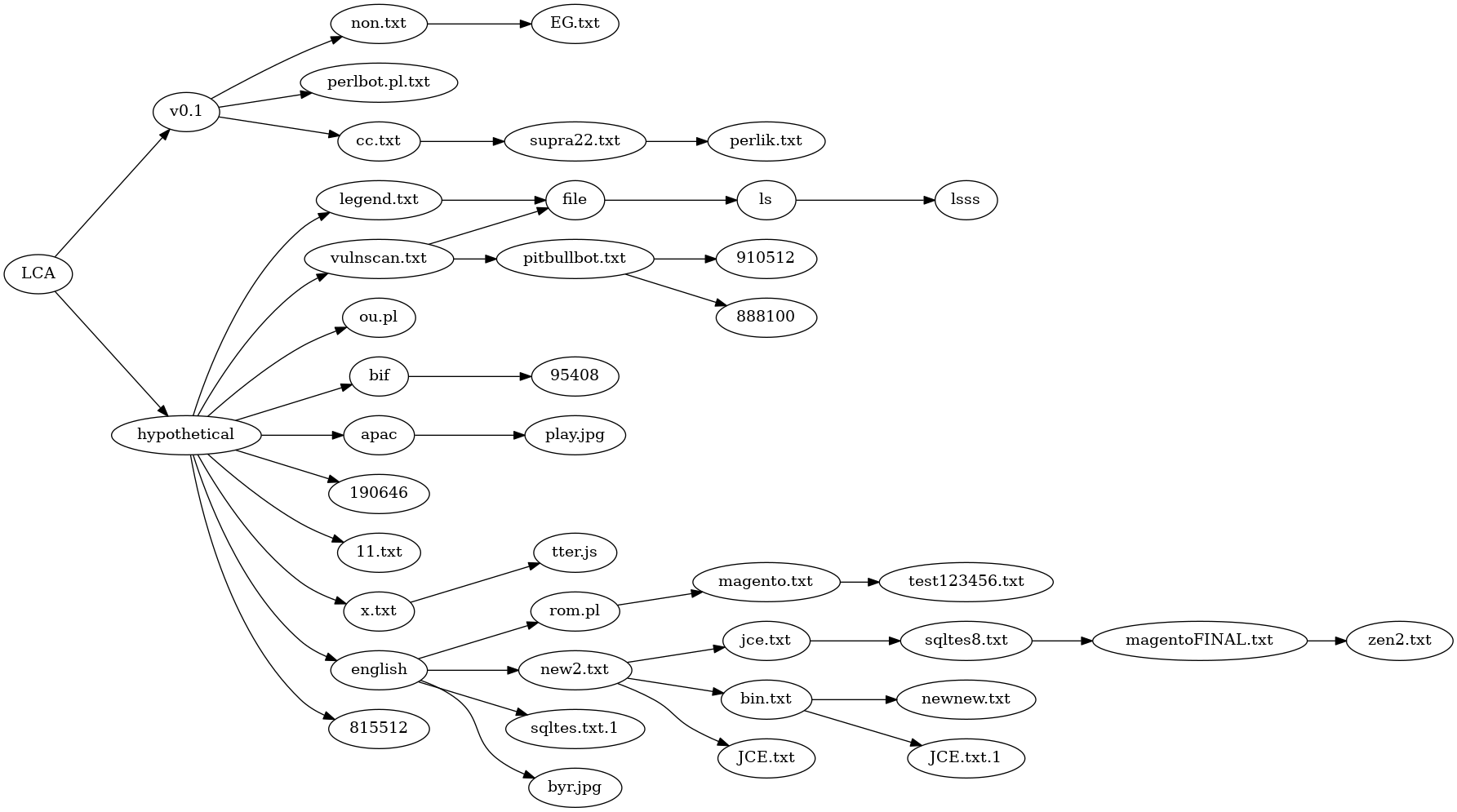
Above is the best phylogeny I could find, derived from close comparison of the variants and careful trait selection. It's a disjointed mess. It does seperate the English language variants into their own lines of descent, but on the whole the result is unsatisfactory.
It is possible to make a non-disjoint tree phylogeny with the addition of 3 extra species.
The first species is just a Last Common Ancestor. In my view, it was very similar to
the code in v0.1 or perlbot.pl.txt. This doesn't seem too controversial.
The second added species is labeled "english" in the tree below. That would be the first program with function names and comments translated from Portugese to English. Because all the english variants are so similar, this hypothetical species almost certainly existed.
The third added species is labeled "hypothetical". In reality, the oval labled "hypothetical"
may have comprised a line of descent with gradual modifications, but at the very least
that line of descent lost package DCC and had some code worked over to eliminate references to
and use of package DCC. A single reference to a variable $dcc_sel, remains in every
version of Perl/Shellbot, one reason why I consider presence of package DCC as the
primitive condition.
With 3 hypothetical but not unrealistic additional species, the phylogeny becomes a tree.
The number of immediate descendants of hypothetical makes it unrealistic, given the
lines of descent attested to by the existing versions.
The above tree still does not contain the complexity of descent of this code. For example,
a line select( undef, undef, undef, 0.01 ); appears in 18 of the versions, some English-language,
some Portugese, all of the versions at least somewhat derived. This is one instance of a trait
appearing without apparent ancestors, unlike biological "horizontal gene transfer"
and more like "intelligent design".
Perl/Shellbot occurs in even more versions than WSO does. It's only other
competitor is the pBot family of PHP IRC bots. I suspect this is because it's
been around longer than any web shell, and you have to modify the source code
to configure it. Once the botherder has taken the trouble to configure a server
name and master alias, why not go even further? The modifications tend to be in
non-IRC parts of the code, sub sendraw, sub conectar/connector and sub parse
all stay much the same from the most primitive to the most derived
versions.
Malware that has source code available evolves by feature addition and subtraction. An algorithm that reconstructs phylogenies of source code malware must take this evolution into account. Algorithms considering only feature addition won't be able to reconstruct complicated phylogenies of some real world source code malware.