This repository contains several algorithms for cilia segmentation and other support scripts for further experiments. Although Cilia segmentation is 3 label segmentation, we modify the mask to contain only cilia and background. We used three different approaches to solve the problen:
- Optical Flow
- Unet
- Tiramisu
Data are sequences of frame images. There are 325 videos in total each with 100 consecutive frames of a grayscale video of cilia. 211 of them are for training and 114 for testing.
Each training video also has one mask image with 3 labels. Background pixels are labeled 0, cells are labeled 1 and cilia are labeled as 2.
The output is one mask for each testing video with pixels of cilia being labeled as 2. a
Below are instructions for installing and using this package.
You can use the REQUIREMENTS.txt file to setup the environment. The routine is as below:
conda create --name MY_ENV --file REQUIREMENTS.txt
source activate MY_ENV
python3 -m hastings.__main__ <args>
The following arguments are supported by our model:
- model : Specify the model you want to use. Ex: --model="unet"
- mode : Specify if you want to fit the model or predict. Ex: --mode="fit"
- operation : Specify if you want to preprocess Ex: --operation="preprocess"
- image_path : Specify the path of numpy array of prepared image data Ex: --image_path="/home/ubuntu/img.npy"
- mask_path : Specify the path of numpy array of prepared mask data Ex: --mask_path="/home/ubuntu/mask.npy"
- model_path: Specify the path of trained model Ex: --model_path="/home/ubuntu/model.h5"
- save_path: Specify the path where you want to save result (Model while fitting, predictions while predicting, numpy arrays while predicting. The path is only required till the directory. Ex: --save_path="/home/ubuntu/models/"
- file_path : Specify the path to the train.txt or test.txt files which contain the hash values of folders/ Ex: --file_path="/home/ubuntu/data/train.txt"
- data_path : Specify the path to the data folder which contains each hash folder. Ex: --data_path="/home/ubuntu/data/train/"
- num_images: Specify the number of images to consider for training or testing from each hash folder. Ex: --numImages=16
- Testing all modules
./setup.py test
- Testing one module
python -m pytest tests/[file you want to test]
- Testing one function under one module
python -m pytest tests/[file you want to test]::[function name]
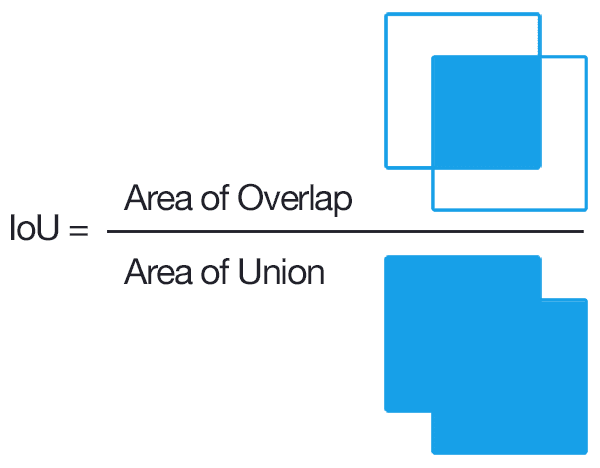
Evaluation of the output masks is done by checking IoU. An illustration for calculating IoU is show asa below.
| Algorithm | Parameter value | IoU |
|---|---|---|
| Optical Flow | threshold = 110 | 16.02 |
| Optical Flow | threshold = 100 | 17.73 |
| Optical Flow | threshold = 90 | 18.97 |
| Optical Flow | threshold = 85 | 19.34 |
| Optical Flow | threshold = 81 | 19.48142 |
| Optical Flow | threshold = 80 | 19.48875 |
| Optical Flow | threshold = 79 | 19.48135 |
| Optical Flow | threshold = 78 | 19.47 |
| Optical Flow | threshold = 75 | 19.40 |
| Optical Flow | threshold = 70 | 19.00 |
The score below is based on the Autolab grader used by Dr.Shannon Quinn for the course.
- optical Flow : 19.48875
- Unet : 4.6
- Tiramisu : Couldn't train or test on the whole dataset.
(Ordered alphabetically)
- Ankit Vaghela - ankit-vaghela30
- Vyom Shrivastava - vyom1911
- Weiwen Xu - WeiwenXu21
See CONTRIBUTORS file for more details.
[1] Olaf Ronneberger, Philipp Fischer, Thomas Brox; U-Net: Convolutional Networks for Biomedical Image Segmentation; arXiv:1505.04597
[2] Simon Jégou, Michal Drozdzal, David Vazquez, Adriana Romero, Yoshua Bengio; The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation; arXiv:1611.09326
[3] Aleksander Klibisz, Derek Rose, Matthew Eicholtz, Jay Blundon and Stanislav Zakharenko; Fast, Simple Calcium Imaging Segmentation with Fully Convolutional Networks; https://arxiv.org/pdf/1707.06314.pdf
[4] OpenCV optical flow; https://docs.opencv.org/3.3.1/d7/d8b/tutorial_py_lucas_kanade.html
[5] Jeremy Howard, Fast.ai deep learning tutorial, https://github.com/fastai/courses/blob/master/deeplearning2/tiramisu-keras.ipynb
[6] Team Canady Unet model, https://github.com/dsp-uga/Canady/blob/master/unet.py
This project is licensed under the MIT License - see the LICENSE file for details.