This repository contains the official source code used to produce the results reported in [TBD] paper.
Table of Contents
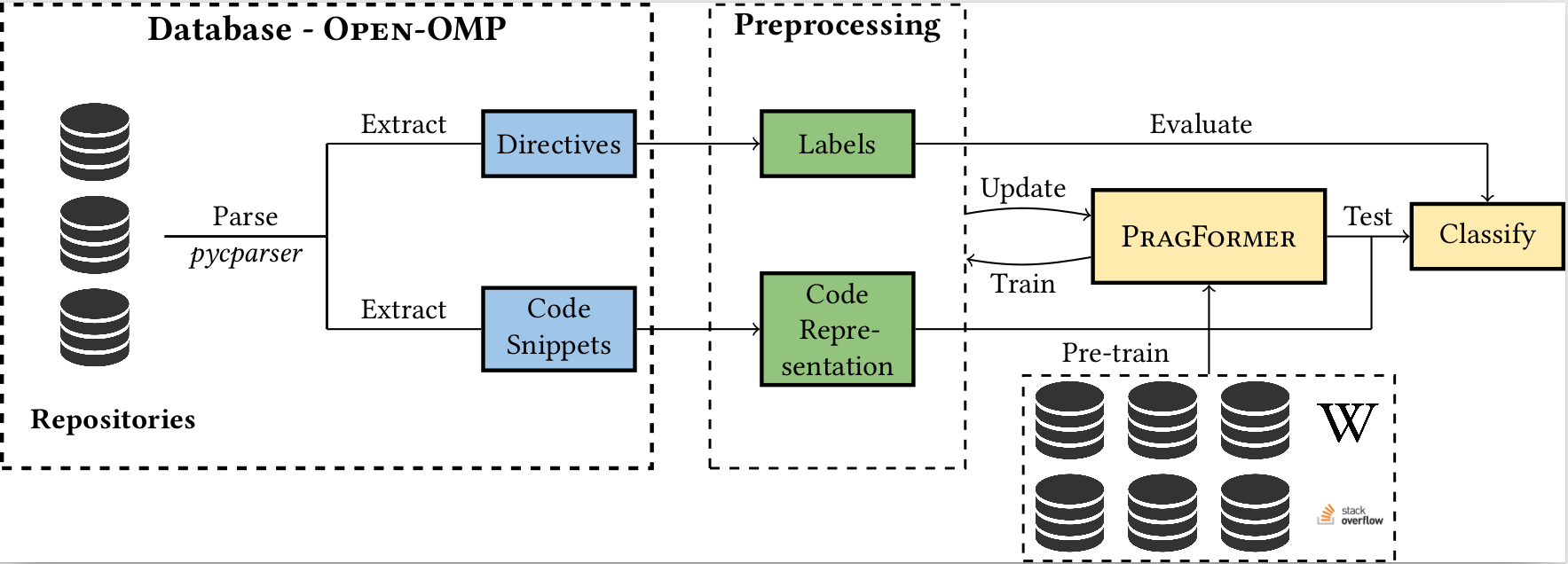
In past years, the world has switched to many-core and multi-core shared memory architectures. As a result, there is a growing need to utilize these architectures by introducing shared memory parallelization schemes to software applications. OpenMP is the most comprehensive API that implements such schemes, characterized by a readable interface. Nevertheless, introducing OpenMP into code, especially legacy code, is challenging due to pervasive pitfalls in management of parallel shared memory. To facilitate the performance of this task, many source-to-source (S2S) compilers have been created over the years, tasked with inserting OpenMP directives into code automatically. In addition to having limited robustness to their input format, these compilers still do not achieve satisfactory coverage and precision in locating parallelizable code and generating appropriate directives. In this work, we propose leveraging recent advances in machine learning techniques, specifically in natural language processing (NLP), to replace S2S compilers altogether. We create a database (corpus), OpenMP-OMP specifically for this goal. OpenMP-OMP contains over 28,000 code snippets, half of which contain OpenMP directives while the other half do not need parallelization at all with high probability. We use the corpus to train systems to automatically classify code segments in need of parallelization, as well as suggest individual OpenMP clauses. We train several transformer models, named PragFormer, for these tasks, and show that they outperform statistically-trained baselines and automatic S2S parallelization compilers in both classifying the overall need for an OpenMP directive and the introduction of private and reduction clauses.
This project is using the following python packages:
pycparser
transformers==3.0.0
pytorch==1.11
matplotlib
huggingface-hub==0.4.0
scikit-learn==0.24.2
scipy==1.7.3
python==3.7
The database is located here It contains folders, whereas each folder contains 3 files:
- code.c - contains the textual code
- pragma.c - contains the pragma of the corresponding code.c -- if no pragma is found, the file doesn't exist
- pickle_code.pkl - contains a class from global_paramerets.py in the project that contains 3 data structures -- the for-loop main node (pycparer node c_ast: For); the pragma node (pycparser node c_ast: Pragma); and the inner function defenitions inside the for-loop (pycparser node c_ast: FuncDef).
Please note, that the pickle file can easily be created by wrapping code.c with a int main() {} and applying the pycparser parser.
In addition, the database contains a json file that maps the folders -- each key in the json file represents a database record: code path, pragma path, pickle path, a unique id and the original file used to create the code (note that is locally, to retrace it simply search for the relevant project and username).
The code used to create the database is located in the PragmaForExtractor in this repository.
This project contains 2 code folders:
- PragmaForExtactor -- contains the code to create the database (Open-OMP.tar.gz) and a necesarry globals_parameter.py to extract the pickle file
- Model -- contains the files to reproduce the paper.
Inside Model there are several files:
- data_creator.py -- creates the pickle file for the model. of course you can create your own pickle file as long as it saved as the Data class inside global_parameters.py (9 fields - train/valid/test and for each, data/label/id)
- train.py -- the training algorithm
- tokenizer.py -- the tokenizer of the code
- predict.py -- prediction algorithm - note that it creates two txt files
- statistics.py -- plot the result or extract number of tokens
- model.py -- define the model (PT-DeepSCC + FC layer)
- global_parameters.py -- defines the data structure for the model
- database_manipulator.py -- formatted access to the database
- main.py -- train/predict the model