Home
Phinch is an open-source framework for visualizing biological data, funded by a grant from the Alfred P. Sloan foundation. This project represents an interdisciplinary collaboration between Pitch Interactive, a data visualization studio in Berkeley, CA, and biological researchers at UC Davis. Whether it's genes, proteins, or microbial species, Phinch provides an interactive visualization tool that allows users to explore and manipulate large biological datasets.
Phinch is optimized for use in the Chrome browser. It currently supports downstream analyses of BIOM 1.0 files Biological Observation Matrix, a JSON-formatted file type (.biom) typically used to represent diverse types of genomic data. Any type of tab-delimited file containing biological data can be converted into a BIOM file (see Quick Start Guide below). All sample metadata and taxonomy/ontology information MUST be embedded in the BIOM file before being uploaded. Note: Phinch does NOT support BIOM 2.0 files or higher (HDF5 format, the new default output format for QIIME 1.9 and higher); these files MUST be converted to JSON-formatted BIOM 1.0 files before they can be loaded into Phinch.
- Quick Start Guide - how to prepare your data
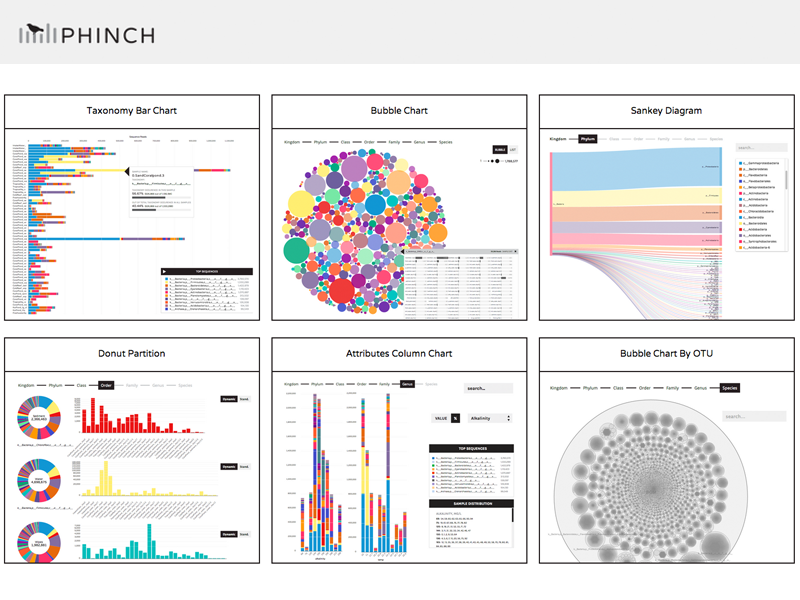
- Visualization Gallery - example screenshots
- Tutorials
- Format - Guide for formatting your sample metadata before embedding it in the .biom file
- Talks
- People
- Release Notes
- Phinch Google Group - post here to request software support, ask questions, report bugs and errors, etc.
Our draft manuscript is currently available as a preprint on bioRxiv. Please cite Phinch as follows:
Bik, H.M. and Pitch Interactive (2014) Phinch: An interactive, exploratory data visualization framework for –Omic datasets, bioRxiv, doi: http://dx.doi.org/10.1101/009944
Phinch supports so-called “modern” browsers, which generally means everything except IE8 and below. D3 is tested against Firefox, Chrome (Chromium), Safari (WebKit), and Opera. Parts of Phinch may work in older browsers. You'll need a modern browser to use SVG and CSS3 Transitions.