PLAIT: Phenotyping Lymphoma with an Artificial Intelligence Toolbox
The anonymization app deletes personal data from .fcs/.lmd files. In the anonymization process, the entered file is overwritten in the affected areas, which means field contents are deleted. This enables the further use of the anonymized file for physicians and doctors on services such as ours.
Requirements: R > 3.5, various shiny-related packages (see below) and RTools
Recommended: R 4.0.3, R-Studio 1.33 and RTools40
PLAIT-Website: https://plait.uni-marburg.de (eduroam VPN necessary)
PLAIT-Webservice: https://plait-ai.uni-marburg.de (eduroam VPN necessary)
Note: For detailed installation instructions go to https://plait.uni-marburg.de/anonymisierungsapp
Install the required packages for the execution (see here; link is accessible from eduroam only - use VPN):
requiredPackages <- c("shiny", "shinyjs", "shinyFiles", "shinyFeedback", "Rcpp")
new.packages <- requiredPackages[!(requiredPackages %in% installed.packages()[,"Package"])]
if(length(new.packages)) install.packages(new.packages, quiet=TRUE)
invisible(lapply(requiredPackages, library, character.only = TRUE))
Run the AnonymizationTool in R with following Command:
shiny::runGitHub("AnonymizationTool", "PLAIT-project", subdir = "R")
For permanent installation, download this repository as ZIP-folder (upper right, green button) and install it locally (make sure that the package is unpacked).
Click on "AnonymizationTool" for opening the Project in R.
To start the app, enter the following command in the command line:
shiny::runApp('R')
More detailed instructions can be found in the Manual.
The full manual for physicians, doctors and scientists who have .fcs/.lmds files for anonymization, is available here; link is accessible from eduroam only - use VPN)
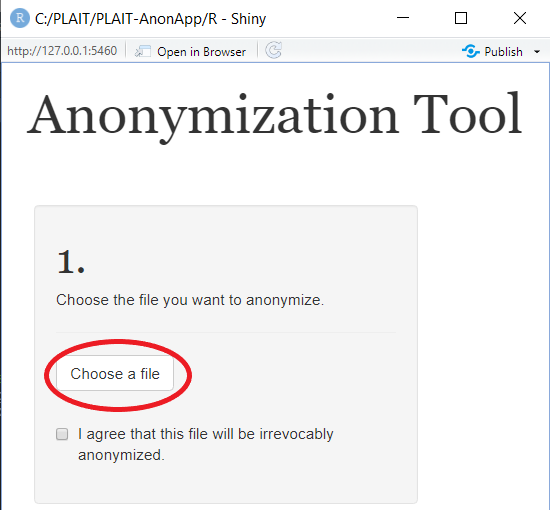
After download, installation and execution, a new window should pop-up:
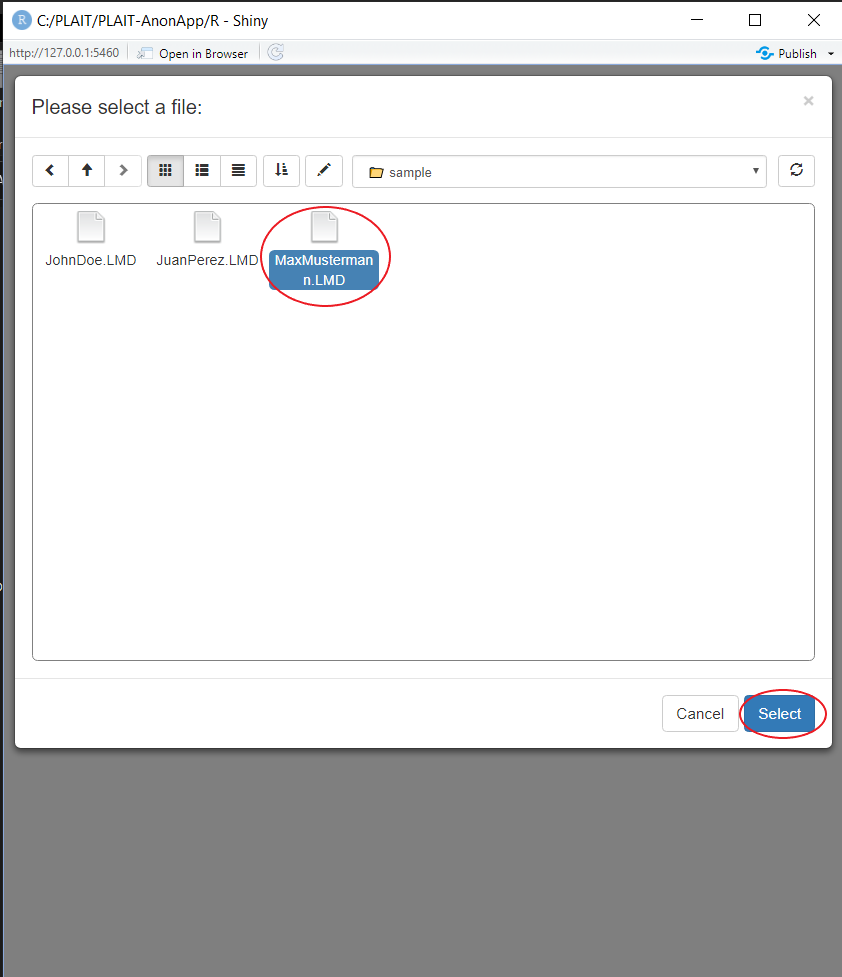
Click on the Button "Choose File", this will prompt your file explorer. Choose the file you would like to anonymize by marking and selecting it. If you'd like to try it out, this package comes with three sample .LMD files in the folder /Sample/.
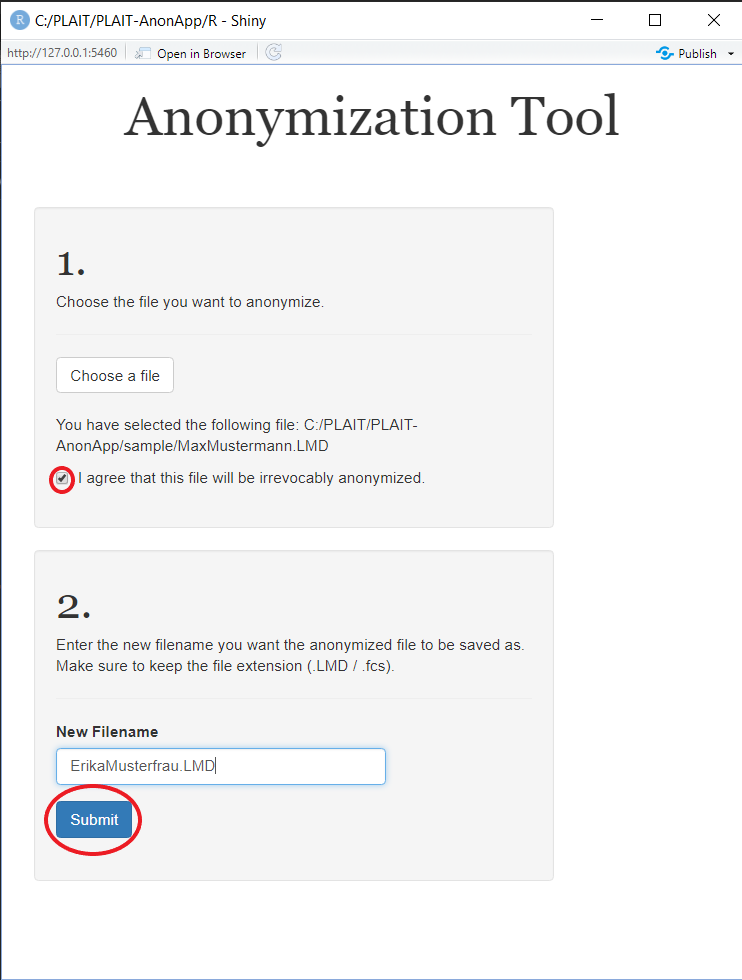
After you have selected your lymphoma dataset (.LMD/.FCS), confirm your wish to irreversibly anonymize this file. It may be recommended, creating a backup of you file in another location of your computer's directory. To avoid this, give your file another name with the same file type extension.
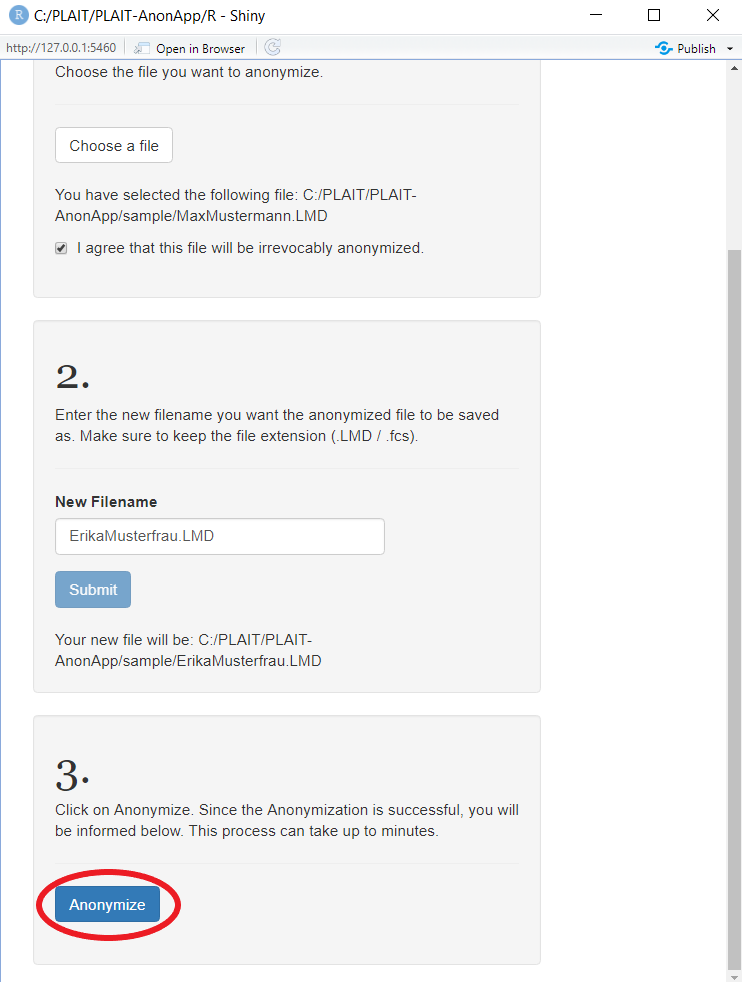
Now, you are all set to let the Anonymization tool do its work: Click on the button "Anonymize"
Note that this is a student project!
For inquiries and feedback, contact: databionics@informatik.uni-marburg.de
Project lead: Prof. Ultsch, Dr. rer. nat. MC Thrun, Dr. med. Brendel
Authors and contributors: Dr. F. Lerch, J. Schulz-Marner, AC. Rathert, C. Kujath, S. Nguyen