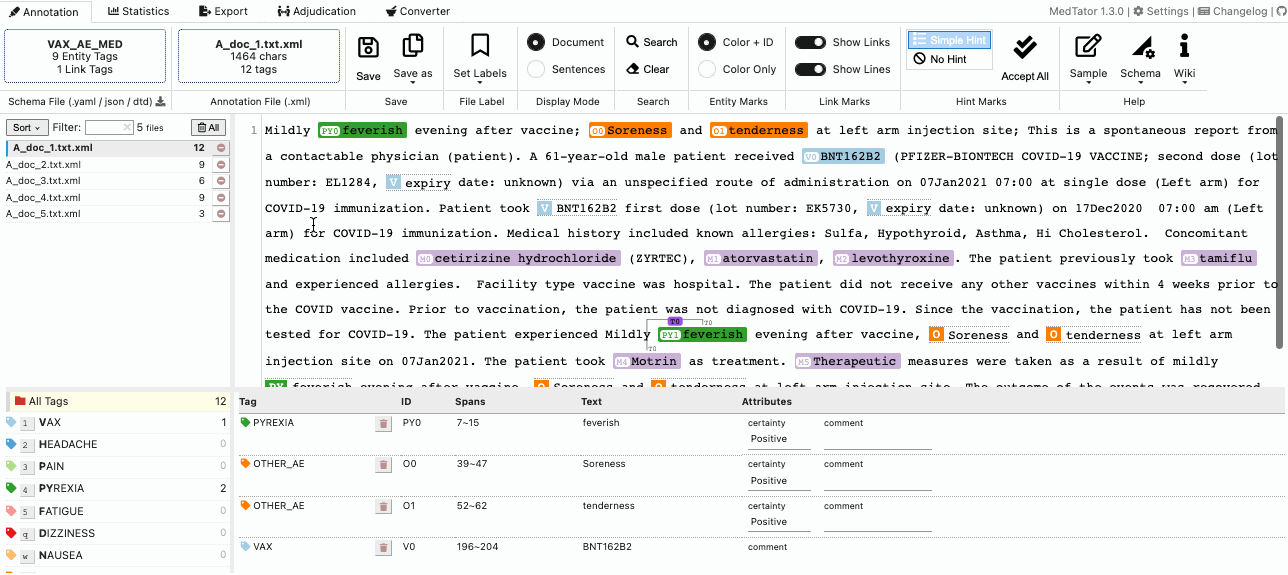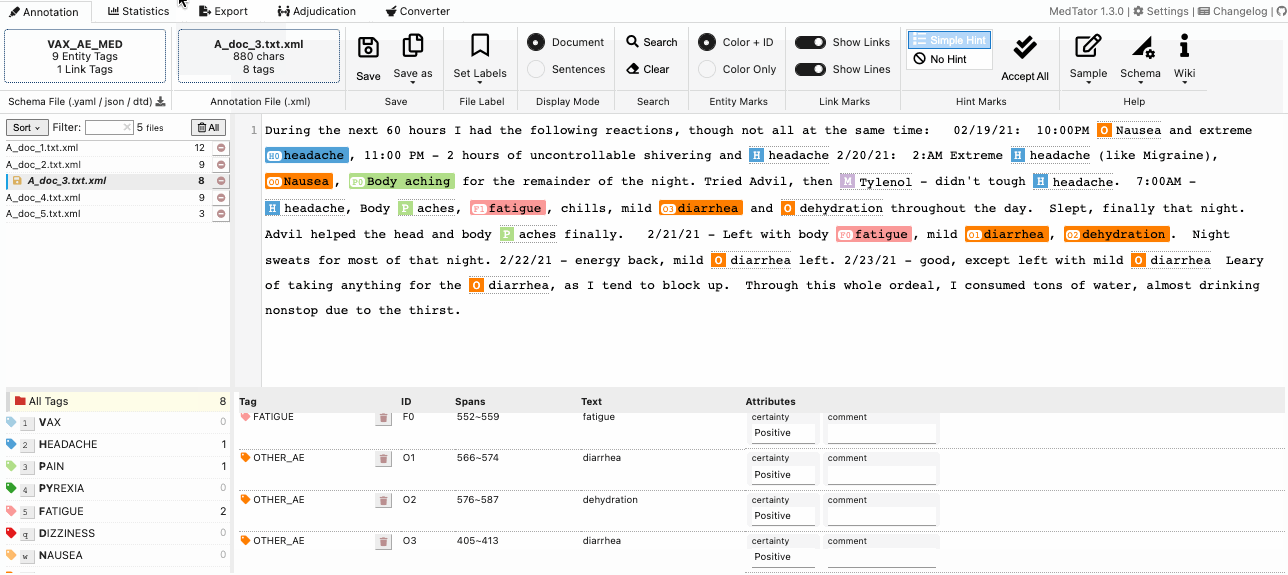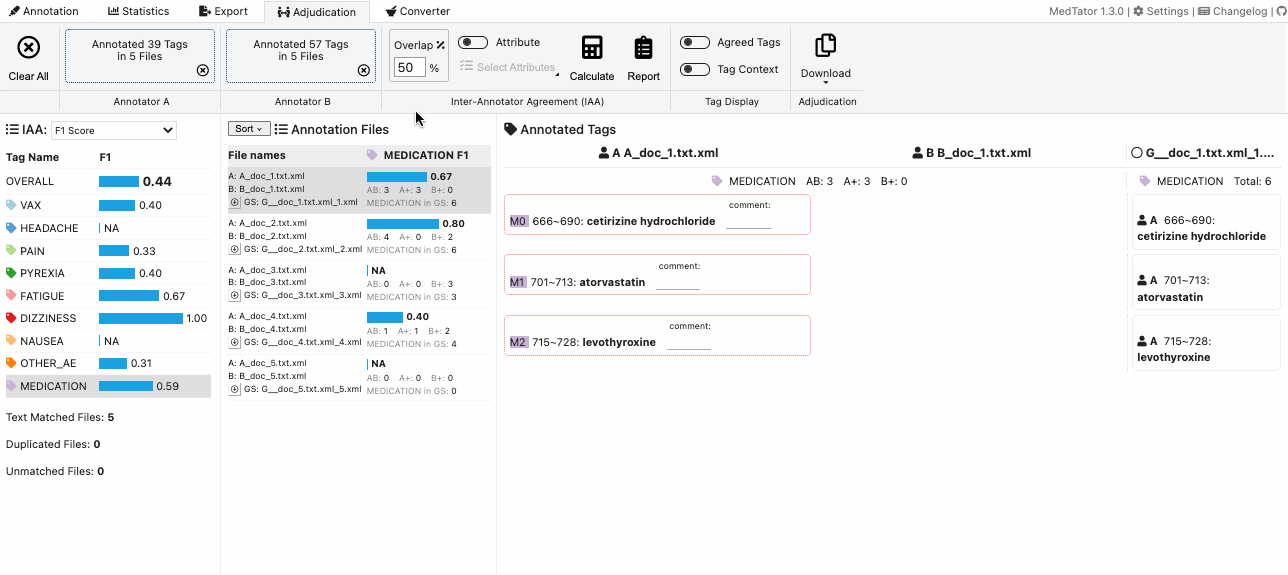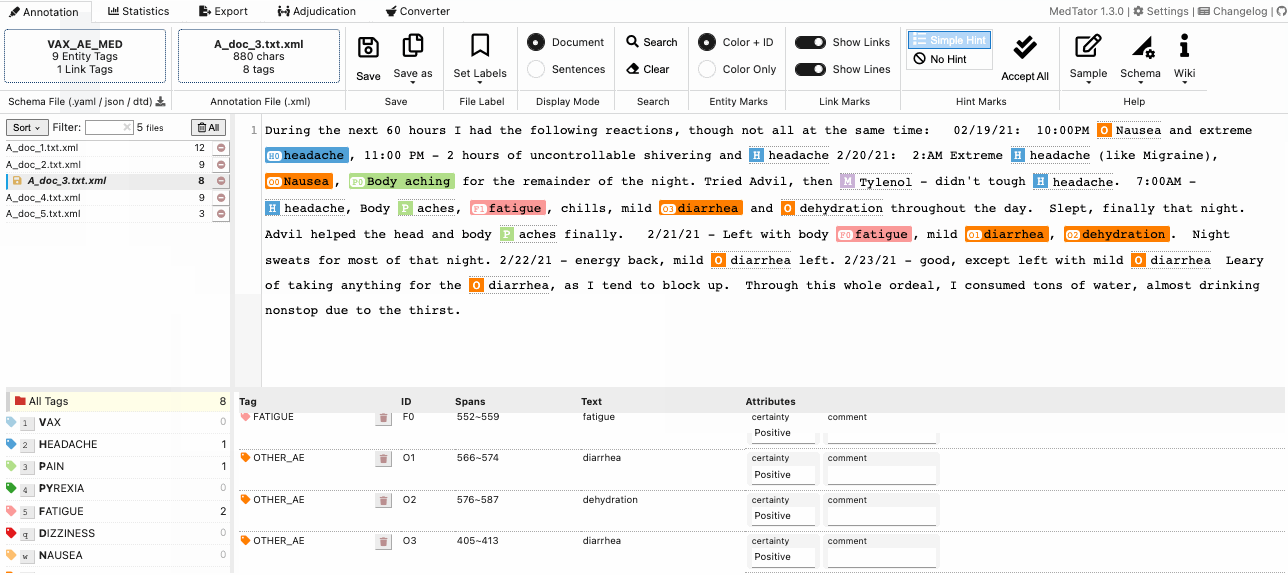
MedTator is a serverless text annotation tool for corpus development. It is built on HTML5 techniques and many open-source packages, and was designed to be easy-to-use for your annotation task.
No Java, no Python, no PHP, no Docker, no MySQL, and no need to install any server or client runtime for corpus annotation! Check Here to Start Annotation Now!
If you're having trouble using MedTator, you can use Issues to tell us about the issue you're experiencing.
- New to MedTator? Let's have a Quick Start.
- More samples? Check our Sample Datasets.
- No internet access? Check this Standalone version
- Effective strategies for annotating? Check this Annotation best practices
- Design annotation schema? Check this Annotation Schema Design.
- Parse annotation data? Here is the Annotation File Format.
- Use annotated files for machine learning? We have some Python Sample Scripts.
- Having issues? Check the Issues.
- More details about MedTator? We have a Manual.
- And more contents in the Wiki
MedTator itself doesn't require Python runtime environment, so you don't need to install any runtime environment to run MedTator for corpus annotation. If you are interested in the MedTator development or just want to try the development version, a Python 3+ runtime environment is needed to run a debugging server.
You can install a Python 3+ or Miniconda / Anaconda, then download the source code of MedTator and install the requirements (just Python Flask, that's all):
pip install -r requirements.txtThen, run the following command to start a local server which is binding port 8086:
python web.pyNow you can open web browser and check the http://localhost:8086/.
For more details of the parameters for web.py, run python web.py -h and it will show the details as follows.
usage: web.py [-h] [--mode {build,run,release}] [--lib {local,cdn}]
[--path PATH] [--fn FN]
MedTator Development Server and Toolkit
optional arguments:
-h, --help show this help message and exit
--mode {build,run,release}
What do you want to do? `run` for starting the
development server. `build` for generating a static
HTML page for public release or local release.
--lib {local,cdn} Where to get third party libs in the HTML page? If
choose local, please make sure to copy the `static`
folder after generated the HTML file.
--path PATH Which folder to be used for the output page? The
default folder is the docs/ folder for public release.
--fn FN What file name to be used for the output page? The
default file name is the `index.html` which could be
accessed directly by browser.
To update the static version for publication (e.g., GitHub Pages), run the following command. It will generate a static HTML file in the docs/ folder and copy other files.
In addition, as the default filename is index.html, the build script will automatically create a index.VERSION.html in the build path for backup.
So that the user can access the old version for comparison or checking old functions.
python web.py --mode buildOr you can build a dev version for public testing, run the following command:
python web.py --mode build --fn dev.htmlOr you can build a standalone version for local use, run the following command:
python web.py --mode build --lib local --fn standalone.htmlThen, you can create a release zip file:
python web.py --mode releaseApache-2.0 License
If you use MedTator in scientific work or want to learn more about it, please take a look at our paper:
He H, Fu S, Wang L, Liu S, Wen A, Liu H. MedTator: a serverless annotation tool for corpus development. Bioinformatics, Volume 38, Issue 6, 15 March 2022, Pages 1776–1778, DOI: 10.1093/bioinformatics/btab880, PMID: 34983060
- Research on IOB2/BIO format editing
- Research on UMAP algorithm
- Research on in-browser text embedding
- Add sample dataset for error analysis
- Update icon for error analysis
- Update Cohen's Kappa calculation
- Update README for new features
- Update documents for error analysis
- Update documents for new schema
- Update UI for error analysis
- Update demo script for inter-sentence
- Added example Python script for masking entities
- Updated medtator_kits.py for saving xml
- Updated relation annotation example schema and data
- Fixed one-offset bug in first-line newline sign
- Fixed cross-line entity render bug
- Added workspace JSON drag and drop on file list
- Added workspace saving function
- Added workspace loading function
- Added schema drag and drop on file list
- Added shortcut ALT + ↑ / ↓ to move to prev / next file
- Added Toolkit/MedTaggerVis for checking .ann files (experimental)
- Added local setting cache for auto save/load (experimental)
- Fixed error analysis Sankey link bug
- Fixed error analysis SVG height bug
- Fixed error analysis popup box bug
- Updated design for .ann/.txt conversion
- Updated FAQ in MedTator Wiki
- Updated scripts for downstream tasks
- Updated sample datasets for new settings
- Added annotation visualization based on brat
- Added Python scripts for read/parse MedTator XML format
- Added Jupyter notebook for downstream tasks with MedTator XML files.
- Developed functions for brat vis format conversion
- Developed tag extraction based offset spans
- Updated style for link/relation display
- Added schema of .yml extension support
- Added auto-save (experimental) feature
- Fixed scheme editor open bug
- Update the UI for IAA
- Updated a data sample for error analysis
- Added label panel for error labeling
- Added error sankey diagram for error analysis
- Added token scatter for error analysis
- Added heatmap of error distribution for error analysis
- Added bar charts of error statistics for error analysis
- Added tag list and menu items for error analysis
- Updated scripts for text embedding web services
- Fixed IOB2/BIO empty export bug
- Fixed exporter window height bug
- Fixed BioC format export annotation text encoding bug
- Fixed BioC format export attribute text encoding bug
- Added functions for error analysis
- Added Math.js for statistics and matrix
- Added ECharts for visualization
- Added a sample dataset for error analysis
- Updated the UI design for adjudication tab
- Updated the packaged used to reduce loading time
- Fixed open annotation files dialog bug
- Fixed IAA download bug
- Refactored file reading workflow with asynchronous processing
- Refactored annotation file loading workflow
- Refactored code structure to reduce code size
- Designed JSON/YAML format schema
- Added tokenization exceptions
- Added pagination for large dataset
- Added loading dataset annimation
- Added drag and drop folder for adjudication
- Added drag and drop folder for converter
- Added converting text files to MedTator xml
- Added a float text content viewer
- Added reading JSON/YAML for schema editor
- Added JSON/YAML download option for schema editor
- Added cmd/ctrl + s shortcut key for saving current file
- Updated annotation UI design for bigger drag&drop area
- Updated converter UI design for bigger buttons
- Updated converter for downloading individual result file
- Updated converter with support of viewing contents
- Updated event handlers for drag and drop events
- Updated license files of imported libraries
- Updated the sample schemas with JSON/YAML format
- Added function for customizing sample text
- Added changelog information on UI
- Added sorting for corpus token list in statistics
- Added tag details for corpus summary in statistics
- Added filter options for token statistics
- Added filtered count in token summaryies
- Added a remote corpus sample
- Updated all sample datasets
- Updated the visual design for corpus summary in statistics
- Updated the color encoding for annotated tags in statistics
- Updated scripts for experimental tasks
- Fixed ribbon menu resize bug
- Added meta-data structure for annotation file
- Added color label to annotation file
- Added functions for updating annotator for adjudication
- Added download all tags for adjudication
- Added sorting by label color
- Added MedTagger ann format converter
- Added UI for converting MedTagger results
- Added folder drag and drop for converter
- Added folder drag and drop for adjudication
- Updated IAA drop box width
- Updated code structure for better extension
- Updated wiki pages
- Fixed meta copy bug when generating adjudicated copy
- Added sample text for schema test
- Added Cohen's Kappa score to exported XLSX report
- Updated texts on UI
- Updated CDN URLs for some packages
- Fixed default IAA sample data bug
- Added seperate file download for adjudicated file
- Added annotator label for adjudicated files
- Updated adjudication tab UI
- Fixed edit bug
- Added the sort for IAA file list
- Updated iaa result box style
- Updated sample dataset
- Fixed duplicated tag in adjudicated results
- Fixed ajax request type bug
- Fixed adjudicated result download bug
- Added sample schema dropdown for editor to select
- Added sample schema files for standalone version
- Added help to schema editor with wiki
- Updated the tooltip information for annotation menu
- Updated scripts for creating sample DTD files
- Fixed the initial position of context menus
- Fixed schema editor menu UI margin
- Fixed path missing bug in the export script
- Added Cohen's Kappa to IAA calculation results
- Added dropdown for selecting Cohen's Kappa for IAA result
- Added confusion matrix for Cohen's Kappa
- Added schema editor for creating and editing schema
- Added helper functions for building schema
- Added schema editor convert functions
- Updated annotation bar menu for schema editor
- Updated dtd stringify function
- Updated sample data for demo
- Updated browser detection for Brave
- Updated the size of search button
- Fixed the message for secure contexts
- Fixed the message for browsers without FSA APIs
- Added sort button and menu for file list by file name or tags
- Added dynamic sort indicator for different sort mode
- Added a button for quickly deleting filter text
- Fixed concept name overflow bug in adjudication tab
- Fixed ribbon menu layout bug when resized
- Added using attribute IAA calculation
- Added menu for attribute selection
- Added dropping file in the filelist view
- Updated the drop zone size
- Updated the schema and IAA sample dataset
- Updated the the JSON files of the samples
- Fixed switch button status when no dtd file
- Added a new section in the statistics tab
- Added a new statistics report in xlsx format
- Added colored cells to the statistics report
- Added importing annotation files by dropping folder(s)
- Added memory check in settings
- Updated the statistics summary with more items
- Updated the XML zip file name with timestamp
- Added the adjudication sheet in the IAA report xlsx file
- Updated the included tags in the IAA report xlsx file
- Updated the colors in IAA report xlsx file
- Updated wiki pages
- Added a setting option for showing annotated tags
- Added fixed header for the "All Tags" in concept list
- Updated the context menu position calculation
- Fixed the hover bug dismiss bug when deleting tag
- Added IAA report download (as Excel XLSX format)
- Added background colors to IAA report cells
- Added summary and files details in the IAA report
- Updated Wiki button for issue report
- Updated IAA sample data annotation and schema design
- Updated sample json files for style information
- Updated the document-level adjudication label
- Updated default samples
- Fixed "Clear All" button not working bug
- Added hover box to entity tag for details
- Added show adjudication in annotation tab with
- Added download all annotation as a zip file
- Added fixed tag list header scrolling
- Added download schema dtd file
- Added dynamic tag rendering based on selected tag
- Added filter hinter based on selected tag
- Added clear seperate adjudication
- Updated the export message link
- Updated messages related to annotation file import
- Updated messages related to dtd file import
- Updated simple sentencize algorithm
- Updated sentencizer algorithm
- Updated adjudication tab design
- Updated mark style in editor
- Updated hover text style for tag and hint
- Updated animated tag style in tagging view
- Updated active style for tag list
- Updated conditions for saving annotation file
- Updated sample datasets
- Fixed rendering glitch when switching tab
- Fixed IAA calculation click not binding bug
- Fixed sentencizer setting not working bug
- Fixed document-level tag rendering bug
- Fixed tag attribute toggle in adjudication
- Fixed popup menu dobule click
- Fixed popup menu initial position
- Fixed hover tag text shaking bug
- Fixed IAA accept button bug
- Added search and clear search results in editor
- Added highlight in tag list
- Added script for standalone version export
- Updated menu item for exported rulesets
- Updated tool tips
- Updated samples for entity and relation annotation
- Added a new standalone version for local usage without internet access
- Added build parameters for building standalone version
- Added local cache of all libraries for local standalone version
- Updated references configuration for standalone version
- Updated embedded sample data for standalone version
- Updated building output information
- Fixed drag & drop bug when reading local dtd file
- Fixed embedded sample JSON encoding bug
- Added a sentence splitting / tokenization method for fast splitting.
- Added a side bar for managing settings.
- Added entity tag locating. Users could click on the
spansto locate the entity tag in the editor. The editor will jump to the line where the clicked tag is and highlight that tag.
- Added sample data for AMIA 2021 workshop.
- Fixed typos.
Release Highlights
- Serverless design. NO runtime installation is required.
- Four tabs for core steps in annotation.
- Visualized entity and relation annotation.
- Annotation hint based on existed tags.
- Visualized annotation results.
- Interactive adjudication and Inter-Annotator Agreement (IAA) calculation.
- Customizable annotation schema.
- Multi-format export.
- Multi-file annotation.
We thank all of our team members for their insights and contributions.