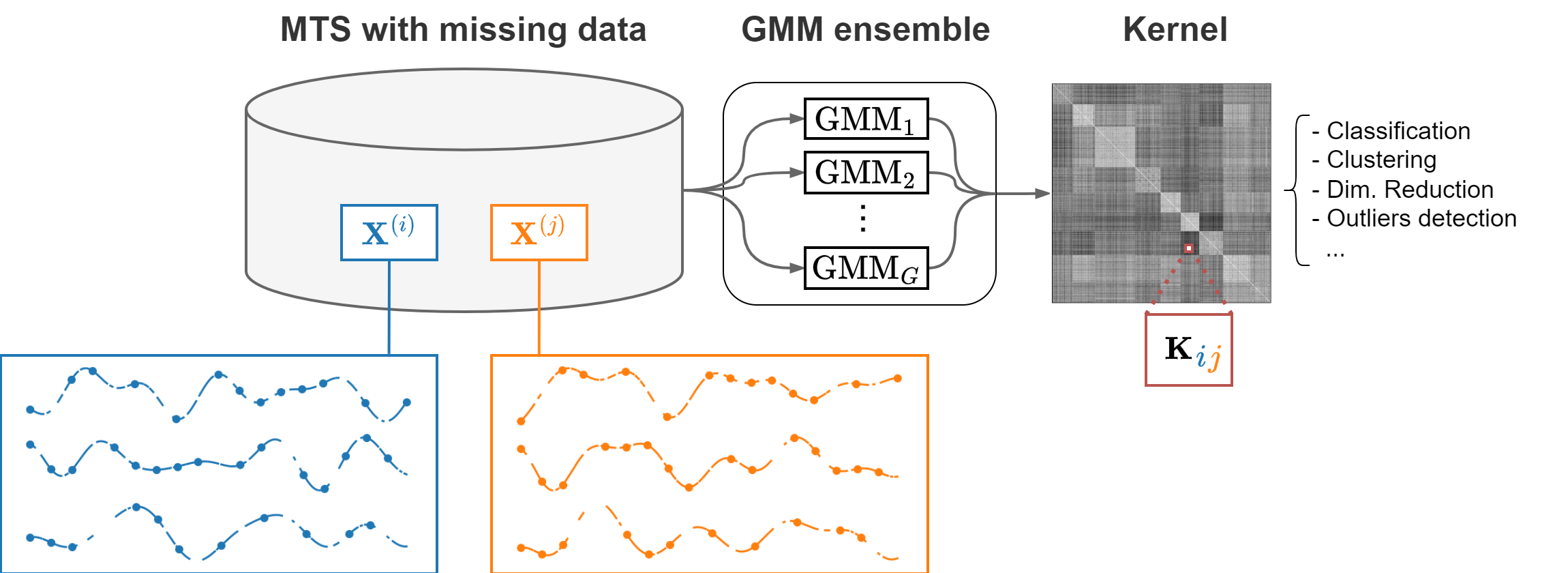
The Time Series Cluster Kernel (TCK) is a kernel similarity for multivariate time series with missing values. Once computed, the kernel can be used to perform tasks such as classification, clustering, and dimensionality reduction.
TCK is based on an ensemble of Gaussian Mixture Models (GMMs) for time series. The GMMs use time-varying means to handle time dependencies and informative Bayesian priors to handle missing values. The similarity between two time series is proportional to the number of times they are assigned to the same mixtures.
The recommended installation is with pip:
pip install tckAlternatively, you can install the library from source:
git clone https://github.com/FilippoMB/https://github.com/FilippoMB/Time-Series-Cluster-Kernel.git
cd https://github.com/FilippoMB/Time-Series-Cluster-Kernel
pip install -e .The following scripts provide minimalistic examples that illustrate how to use the library for different tasks.
To run them, download the project and cd to the root folder:
git clone https://github.com/FilippoMB/https://github.com/FilippoMB/Time-Series-Cluster-Kernel.git
cd https://github.com/FilippoMB/Time-Series-Cluster-KernelClassification
python examples/classification.pyClustering
python examples/clustering.pyThe following notebooks illustrate more advanced use-cases.
TCK uses multiprocessing. While using multiprocessing in Python on Windows, it is necessary to protect the entry point of the program by using
if __name__ == '__main__':Please, refer to the following examples.
Classification
python examples/classification_windows.pyClustering
python examples/clustering_windows.pyData format
- TCK works both with univariate and multivariate time series. The dataset must be stored in a numpy array of shape
[N, T, V], whereNis the number of variables,Tis the number of time steps, andVis the number of variables (V=1in the univariate case). - If the time series in the same dataset have a different number of time steps,
Tcorresponds to the maximum length of the time series in the dataset. All the time series shorter thanTshould be padded with trailing zeros to match the dimensionT. Alternatively, one can use interpolation to stretch the shorter time series up to lengthT. - The time series can contain missing data. Missing data dare indicated by entries
np.nanin the data array.
Available datasets
There are several univariate and multivariate time series classification datasets immediately available for test and benchmarking purposes.
To list of available datasets can be retrieved as follows
from tck.datasets import DataLoader
downloader.available_datasets(details=True) # Leave at False to just get the namesA dataset can be loaded as follows
Xtr, Ytr, Xte, Yte = downloader.get_data('Japanese_Vowels')There are few hyperparameters that can be tuned to modify the TCK behavior.
tck = TCK(G, C)Gis the number of GMMs.Cis the number of components in the GMMs.
Usually, the higher the better but the computations take longer.
tck.fit(X, minN, minV, maxV, minT, maxT, I)minN: Minimum percentage of samples to be used in the training of the GMMs.
minV: Minimum number of attributes to be sampled from the dataset.
maxV: Maximum number of attributes to be sampled from the dataset.
minT: Minimum length of time segments to be sampled from the dataset.
maxT: Maximum length of time segments to be sampled from the dataset.
I: Number of iterations for the MAP-EM algorithm.
These parameters are usually less sensitive and can be left to their default value in most cases.
Ktr = tck.predict(mode='tr-tr')
Kte = tck.predict(Xte=Xte, mode='tr-te')- If
mode='tr-tr', returns the similarity matrix between training samples, i.e.,Ktr[i,j]is the similarity between time seriesiandjin the training set. - If
mode='tr-te', it is necessary to pass the test setXteas additional imput. The returned similarity matrixKte[i,j]is the similarity between time seriesiin the test set and time seriesjin the training set.
Please, consider citing the original paper if you are using TCK in your reasearch.
@article{mikalsen2018time,
title={Time series cluster kernel for learning similarities between multivariate time series with missing data},
author={Mikalsen, Karl {\O}yvind and Bianchi, Filippo Maria and Soguero-Ruiz, Cristina and Jenssen, Robert},
journal={Pattern Recognition},
volume={76},
pages={569--581},
year={2018},
publisher={Elsevier}
}