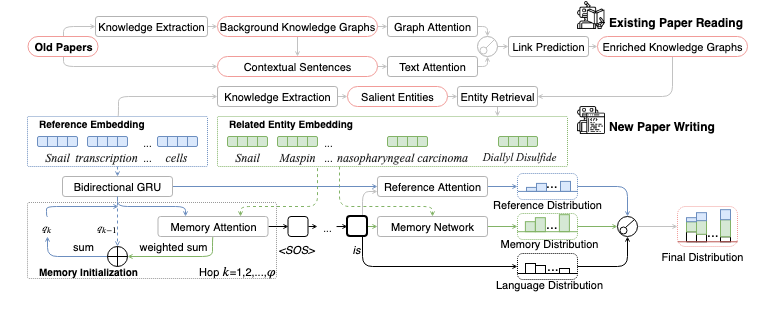
PaperRobot: Incremental Draft Generation of Scientific Ideas [Sample Output]
Accepted by 57th Annual Meeting of the Association for Computational Linguistics (ACL 2019)
- Python 3.6 CAUTION!! The model might not be saved and loaded properly under Python 3.5
- Ubuntu 16.04/18.04 CAUTION!! The model might not run properly on windows because windows uses backslashes on the path while Linux/OS X uses forward slashes
You can click the following links for detailed installation instructions.
-
PubMed Paper Reading Dataset This dataset gathers 14,857 entities, 133 relations, and entities corresponding tokenized text from PubMed. It contains 875,698 training pairs, 109,462 development pairs, and 109,462 test pairs.
-
PubMed Term, Abstract, Conclusion, Title Dataset This dataset gathers three types of pairs: Title-to-Abstract (Training: 22,811/Development: 2095/Test: 2095), Abstract-to-Conclusion and Future work (Training: 22,811/Development: 2095/Test: 2095), Conclusion and Future work-to-Title (Training: 15,902/Development: 2095/Test: 2095) from PubMed. Each pair contains a pair of input and output as well as the corresponding terms(from original KB and link prediction results).
CAUTION!! Because the dataset is quite large, the training and evaluation of link prediction model will be pretty slow.
Download and unzip the paper_reading.zip from PubMed Paper Reading Dataset
. Put paper_reading folder under the Existing paper reading folder.
Hyperparameter can be adjusted as follows: For example, if you want to change the number of hidden unit to 6, you can append --hidden 6 after train.py
python train.py
To resume training, you can apply the following command and put the previous model path after the --model
python train.py --cont --model models/GATA/best_dev_model.pth.tar
Put the finished model path after the --model
The test.py will provide the ranking score for the test set.
python test.py --model models/GATA/best_dev_model.pth.tar
Download and unzip the data_pubmed_writing.zip from PubMed Term, Abstract, Conclusion, Title Dataset
. Put data folder under the New paper writing folder.
Put the type of data after the --data_path. For example, if you want to train an abstract model, put data/pubmed_abstract after --data_path.
Put the model directory after the --model_dp
python train.py --data_path data/pubmed_abstract --model_dp abstract_model/
To resume training, you can apply the following command and put the previous model path after the --model
python train.py --data_path data/pubmed_abstract --cont --model abstract_model/memory/best_dev_model.pth.tar
For more other options, please check the code.
Put the finished model path after the --model
The test.py will provide the score for the test set.
python test.py --data_path data/pubmed_abstract --model abstract_model/memory/best_dev_model.pth.tar
Put the finished model path after the --model
The input.py will provide the prediction for customized input.
python input.py --data_path data/pubmed_abstract --model abstract_model/memory/best_dev_model.pth.tar
@inproceedings{wang-etal-2019-paperrobot,
title = "{P}aper{R}obot: Incremental Draft Generation of Scientific Ideas",
author = "Wang, Qingyun and
Huang, Lifu and
Jiang, Zhiying and
Knight, Kevin and
Ji, Heng and
Bansal, Mohit and
Luan, Yi",
booktitle = "Proceedings of the 57th Conference of the Association for Computational Linguistics",
month = jul,
year = "2019",
address = "Florence, Italy",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/P19-1191",
pages = "1980--1991"
}