NLP often relies on the development of a set of gold standard annotations, or ground truth, for the purpose of training, testing and evaluation. Distant supervision (1) is a helpful solution that has given linked data sets a lot of attention in NLP, however the data can be noisy. Human annotators can help to clean up this noise, however for Clinical NLP domain knowledge is usually believed to be required from annotators, making the process for acquiring ground truth more difficult. In addition, current methods for collecting annotation attempt to minimize disagreement between annotators, and therefore fail to model the ambiguity inherent in language. The lack of annotated datasets for training and benchmarking is therefore one of the main challenges of Clinical Natural Language Processing.
We propose the CrowdTruth method for collecting medical ground truth through crowdsourcing, based on the observation that disagreement between annotators can be used to capture ambiguity in text. This repository contains a ground truth corpus for medical relation extraction, acquired with crowdsourcing and processed with CrowdTruth metrics.
This corpus has been referenced in papers:
- Anca Dumitrache, Lora Aroyo, Chris Welty: Crowdsourcing Ground Truth for Medical Relation Extraction. ACM TiiS Vol. 8, Issue 2, 2018.
- Anca Dumitrache, Lora Aroyo, Chris Welty: CrowdTruth Measures for Language Ambiguity: The Case of Medical Relation Extraction. LD4IE at ISWC 2015.
- Anca Dumitrache, Lora Aroyo, Chris Welty: Achieving Expert-Level Annotation Quality with CrowdTruth: The Case of Medical Relation Extraction. BDM2I at ISWC 2015.
|--ground_truth_cause.csv
|--ground_truth_treat.csv
These files contain the processed ground truth for the medical cause and treat relations, in comma-separated format. The columns are:
- SID: unique ID of the sentence and term pair
- relation: medical relation
- sentence: medical sentence
- term1, term2: the 2 medical terms after correction with FactSpan and RelDir; together, they express the relation:
term1 cause of/treated by term2 - b1, b2: the beginning position of each term in the sentence, measured in number of characters
- e1, e2: the end position of each term in the sentence, measured in number of characters
- sentence_relation_score: the sentence relation score of the medical relation; using cosine similarity over the aggregated crowd data, it computes the likelihood that the relation is expressed between the 2 terms in the sentence
- crowd: the score used to train the relation extraction classifier by Chang et al.(4) with crowd data; it is the sentence-relation score, with a threshold to select positive and negative examples equal to 0.5, and rescaled in [0.5, 1] for positives, and [-1, -0.5] for negatives.
- baseline: discrete (positive or negative) labels are given for each data entry by the distant supervision (1) method, based on whether the relation is expressed between the 2 terms in the sentence
- expert: discrete labels based on an expert’s judgment as to whether the baseline label is correct
- test_partition: manual evaluation scores over the sentences where crowd and expert disagreed, used for evaluating the classifier; the sentence-relation score threshold was set at 0.7 for maximum agreement; sentences scored with 0 were determined to be unclear and were removed from testing
- term1_UMLS, term2_UMLS: the original UMLS (4) terms used for distant supervision, before correction with FactSpan and RelDir
- UMLS_seed_relation: the UMLS relation used as a seed in distant supervision to find the given entry
|--ground_truth.csv-metadata.json
Machine-readable description of the ground truth comma-separated files above.
|--/raw
| |--/FactSpan
| |--/RelEx
| |--/RelDir
The raw data collected from crowdsourcing for each of the 3 crowdsourcing tasks.
|--/templates
| |--/FactSpan
| |--/RelEx
| |--/RelDir
The source code of the crowdsourcing tasks, as they were implemented and ran on CrowdFlower.
The dataset used in our experiments contains 3,984 medical sentences extracted from PubMed article abstracts. The sentences were sampled from the set collected by Chang et al.(4) for training the relation extraction model that we are re-using. Chang et al. collected the sentences with distant supervision (1), a method that picks positive sentences from a corpus based on whether known arguments of the seed relation appear together in the sentence (e.g. the treat relation occurs between terms antibiotics and typhus, so find all sentences containing both and repeat this for all pairs of arguments that hold). The MetaMap parser (2) was used to extract medical terms from the corpus, and the UMLS vocabular (3) was used for mapping terms to categories, and relations to term types. The intuition of distant supervision is that since we know the terms are related, and they are in the same sentence, it is more likely that the sentence expresses a relation between them (than just any random sentence).
We started with a set of 8 relations important for clinical decision making, used also by Chang et al., and focused our evaluation effort on the relations cause and treat. These two relations were used as a seed for distant supervision in two thirds of the sentences of our dataset (1,043 sentences for treat, 1,787 for cause). The final third of the sentences were collected using the other 6 relations as seeds, in order to make the data more heterogeneous.
To perform a comparison with expert-annotated data, we randomly sampled a set of 955 sentences from the distant supervision dataset. This set restriction was done not just due to the cost of the experts, but primarily because of their limited time and availability. To collect this data, we employed medical students, in their third year at American universities, that had just taken United States Medical Licensing Examination (USMLE) and were waiting for their results. Each sentence was annotated by exactly one person. The annotation task consisted of deciding whether or not the UMLS seed relation discovered by distant supervision is present in the sentence for the two selected terms.
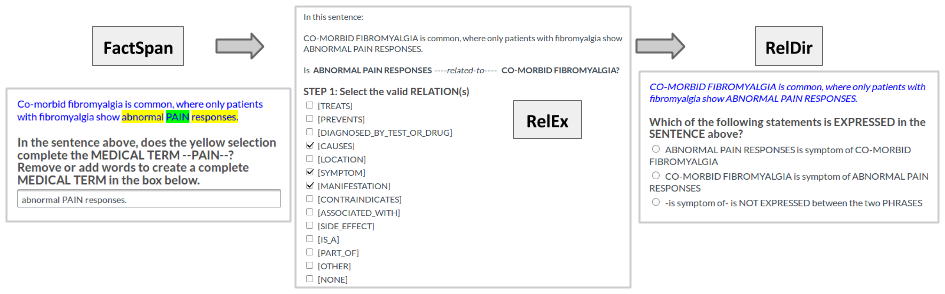
The crowdsourced annotation is performed in a workflow of three tasks. When using terms from the knowledge base to identify candidate sentences, distant supervision does not take into account whether the term is part of a noun phrase -- e.g. distant supervision might identify the term fever as an indicator that a relation is present in a given sentence, when the actual noun phrase in the sentence is viral hemorrhagic fever. Similarly, adjective modifiers (e.g. moderate acute fever) are also not accounted for by distant supervision. To account for this incompleteness, the sentences were pre-processed using a named-entity recognition tool combining the UMLS vocabulary with lexical parsing, to determine whether the seed terms found with distant supervision have alternative word spans. Seed terms that appeared to be incomplete were passed through a crowdsourcing task (FactSpan) in order to get their full word span. In this task, the workers were allowed to add up to 3 words to the left, and three to the right of the seed term, in order to form a complete span. In total, there were 16 possible choices of completing one seed term, including not adding any new words to it. Workers were not allowed to remove words from the seed term, as we did not find this to be an issue in our data.
Next, the sentences with the corrected term spans were sent to a relation extraction task (RelEx), where the crowd was asked to decide which relations hold between the two extracted terms. The workers were able to read the definition of each relation. The task was multiple choice, workers being able to choose more than one relation at the same time. There were also options available for cases when the medical relation was other than the ones we provided (
Finally, the results from RelEx were passed to another crowdsourcing task RelDir to determine the direction of the relation with regards to the two terms. This was a single choice task, and the workers could also select that the relation did not make sense with regards to the given terms.
All three crowdsourcing tasks were run on the CrowdFlower platform with 10-15 workers per sentence, to allow for a distribution of perspectives; the precise settings for each task are available in the table below:
| Task | Workers/Sentence | Payment/Sentence |
|---|---|---|
| FactSpan | 10 | $0.04 |
| RelEx | 15 | $0.05 |
| RelDir | 10 | $0.01 |
The sentences together with the relation annotations were then used to train a manifold model for relation extraction (4). This model was developed for the medical domain, and tested for the relation set that we employ. It is trained per individual relation, by feeding it both positive and negative data. It offers support for both discrete labels, and real values for weighting the confidence of the training data entries, with positive values in (0,1], and negative values in [-1,0).
(1) Mintz, M., Bills, S., Snow, R., Jurafsky, D.: Distant supervision for relation extraction without labeled data. In: Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP: Volume 2. pp. 1003–1011. Association for Computational Linguistics (2009).
(2) Aronson, A.R.: Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. In: Proceedings of the AMIA Symposium. p. 17. American Medical Informatics Association (2001).
(3) Bodenreider, O.: The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research 32(suppl 1), D267–D270 (2004).
(4) Wang, C., Fan, J.: Medical relation extraction with manifold models. In: 52nd Annual Meeting of the ACL, vol. 1. pp. 828–838. ACL (2014).