Genetic algorithm "library"
Current version: 0.2
The "library" was created for a Czech Technical University's course "Problems and algorithms".
It supports evolving individuals composed of fixed-length binary genomes.
Install the library:
pip install https://github.com/chovanecm/python-genetic-algorithm/archive/master.zip#egg=mchgenalg
Or directly:
python setup.py install
The following example shows using the library to solve a very simple problem of maximizing the number of one-values in a vector.
from mchgenalg import GeneticAlgorithm
import numpy as np
# First, define function that will be used to evaluate the fitness
def fitness_function(genome):
# let's count the number of one-values in the genome
# this will be our fitness
sum = np.sum(genome)
return sum
# Configure the algorithm:
population_size = 10
genome_length = 20
ga = GeneticAlgorithm(fitness_function)
ga.generate_binary_population(size=population_size, genome_length=genome_length)
# How many pairs of individuals should be picked to mate
ga.number_of_pairs = 5
# Selective pressure from interval [1.0, 2.0]
# the lower value, the less will the fitness play role
ga.selective_pressure = 1.5
ga.mutation_rate = 0.1
# If two parents have the same genotype, ignore them and generate TWO random parents
# This helps preventing premature convergence
ga.allow_random_parent = True # default True
# Use single point crossover instead of uniform crossover
ga.single_point_cross_over = False # default False
# Run 1000 iteration of the algorithm
# You can call the method several times and adjust some parameters
# (e.g. number_of_pairs, selective_pressure, mutation_rate,
# allow_random_parent, single_point_cross_over)
ga.run(1000)
best_genome, best_fitness = ga.get_best_genome()
# If you want, you can have a look at the population:
population = ga.population
# and the fitness of each element:
fitness_vector = ga.get_fitness_vector()A single crossover point on both parents' organism strings is selected. All data beyond that point in either organism string is swapped between the two parent organisms. The resulting organisms are the children:
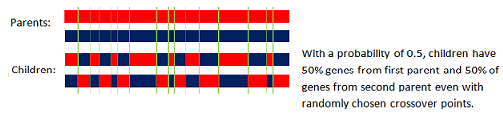
The uniform crossover uses a fixed mixing ratio between two parents. Unlike single- and two-point crossover, the uniform crossover enables the parent chromosomes to contribute the gene level rather than the segment level. If the mixing ratio is 0.5, the offspring has approximately half of the genes from first parent and the other half from second parent, although cross over points can be randomly chosen as seen below:
The mixing ratio in mchgenalg is 0.5.