-
-
Notifications
You must be signed in to change notification settings - Fork 25k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
EHN Optimized CSR-CSR support for Euclidean specializations of PairwiseDistancesReductions
#24556
EHN Optimized CSR-CSR support for Euclidean specializations of PairwiseDistancesReductions
#24556
Conversation
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks you for working on this, @Vincent-Maladiere!
Here are a first few comments to guide you through extending this class hierarchy.
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_radius_neighborhood.pxd.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_radius_neighborhood.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pxd.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pxd.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
Some observations
Testing script:This is working ✅ import numpy as np
from numpy.testing import assert_array_equal
from scipy.sparse import csr_matrix
from sklearn.metrics._pairwise_distances_reduction import ArgKmin
X = np.array([[0, 1], [0, 0], [2, 3]], dtype=np.float64)
Y = np.array([[1, 1], [0, 0], [0, 1]], dtype=np.float64)
X_csr = csr_matrix(X)
Y_csr = csr_matrix(Y)
parameter = 3
dist, indices = ArgKmin.compute(
X,
Y,
parameter,
chunk_size=100,
return_distance=True,
)
dist_csr, indices_csr = ArgKmin.compute(
X_csr,
Y_csr,
parameter,
chunk_size=100,
return_distance=True,
)
assert_array_equal(dist, dist_csr)This fails ❌ (notice that the only difference is the length of X and Y) import numpy as np
from numpy.testing import assert_array_equal
from scipy.sparse import csr_matrix
from sklearn.metrics._pairwise_distances_reduction import ArgKmin
X = np.array([[0, 1], [0, 0], [2, 3]] * 40, dtype=np.float64)
Y = np.array([[1, 1], [0, 0], [0, 1]] * 40, dtype=np.float64)
X_csr = csr_matrix(X)
Y_csr = csr_matrix(Y)
parameter = 3
dist, indices = ArgKmin.compute(
X,
Y,
parameter,
chunk_size=100,
return_distance=True,
)
dist_csr, indices_csr = ArgKmin.compute(
X_csr,
Y_csr,
parameter,
chunk_size=100,
return_distance=True,
)
assert_array_equal(dist, dist_csr) |
Yes, that's right (I supposed you meant "when parameter > Y.shape[0]"?): it's not validated in scikit-learn/sklearn/neighbors/_base.py Lines 786 to 790 in 60cc5b5
As those interface are directly used by users, we do not revalidate all the parameters but this might change in the future. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Nice job making this works, @Vincent-Maladiere!
Here are a few comments to finish this first part.
The next steps involves (in this orders):
- rename
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.{pxd,pyx}.tp(mind the lines mentioning those files as well, IRRC in at least.gitignoreandsetup.cfg) - extending some tests to make sure that the Sparse-Sparse support for Euclidean specialisations is correct
- performing some benchmarks on user-facing API, benchmarking
kneighborsshould suffice (this gist might be adapted) - updating and adapting the documentation/comments of those implementations with respect to those changes
To clarify, the following points better be done as part of subsequent PRs (preferably in this order):
- refactoring for merging of
DatasetsPairsandMiddleTermComputersand encapsulating squared norms computations where appropriate - the support of the CSR-dense and dense-CSR for the Euclidean specialisation
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_gemm_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
|
As most of this work (except |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
As most of this work (except squeuclidean_row_norms) is not user-facing, should we append it to the changelog?
For now, to have the CI green I've labelled this PR with "No Changelog needed" because it is still experimental. Also I converted to to draft because this is still WIP ([WIP] was used before the existence of the draft mode but is not much of an use now). When we have accessed performance changes, we can add an entry to the change log and remove this label.
Also, it looks like a few Cython sources have been added lately.
They should be removable with:
rm sklearn/metrics/_pairwise_distances_reduction/{_gemm_term_computer,_radius_neighborhood}.{pxd,pyx}
git rm --cached -f sklearn/metrics/_pairwise_distances_reduction/{_gemm_term_computer,_radius_neighborhood}.{pxd,pyx}
git add sklearn/metrics/_pairwise_distances_reduction/{_gemm_term_computer,_radius_neighborhood}.{pxd,pyx}
git commit -m "Remove previous Cython templates and sources"
Here are a few comments.
sklearn/metrics/_pairwise_distances_reduction/_middle_term_computer.pxd.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_middle_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
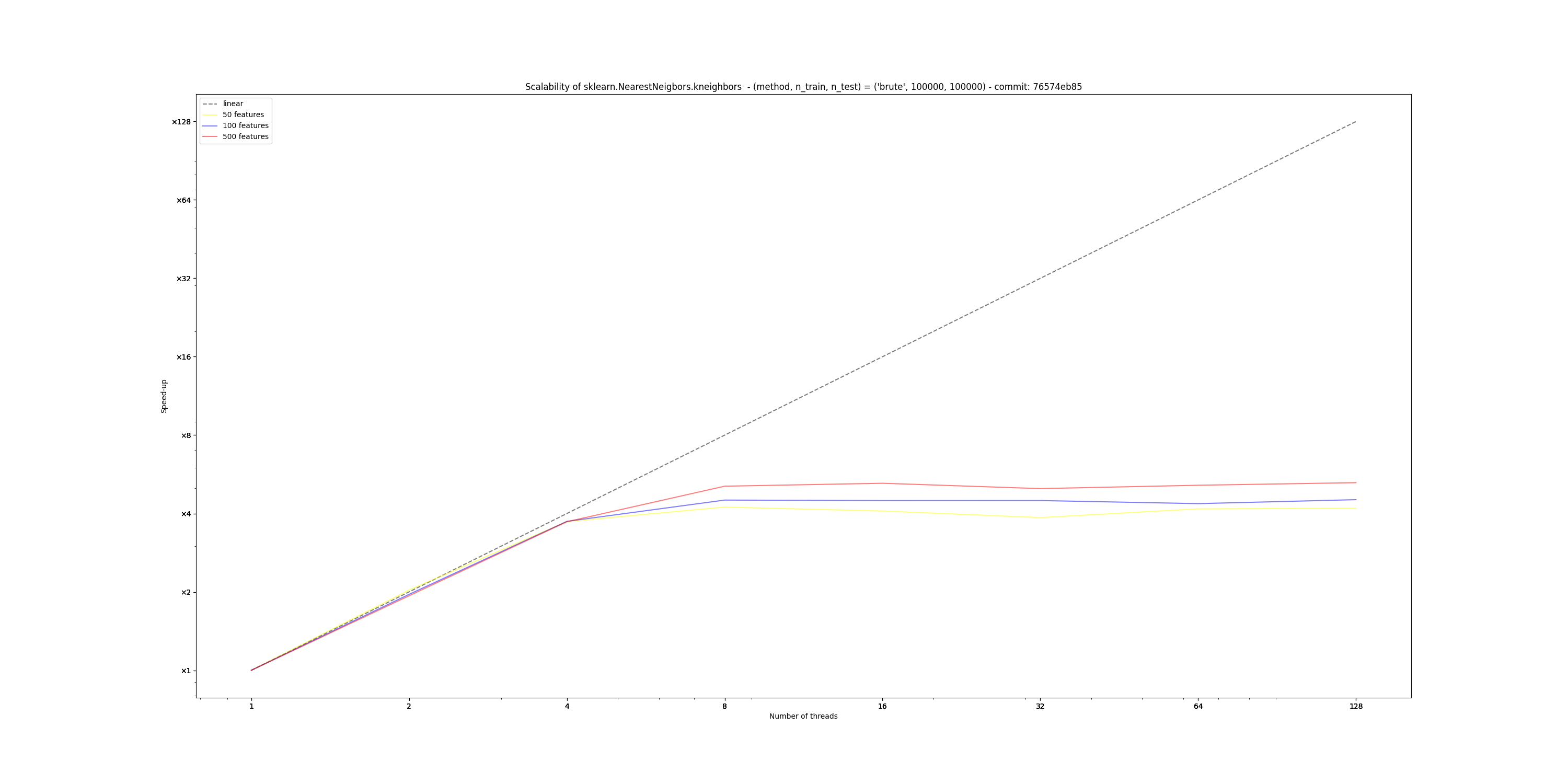
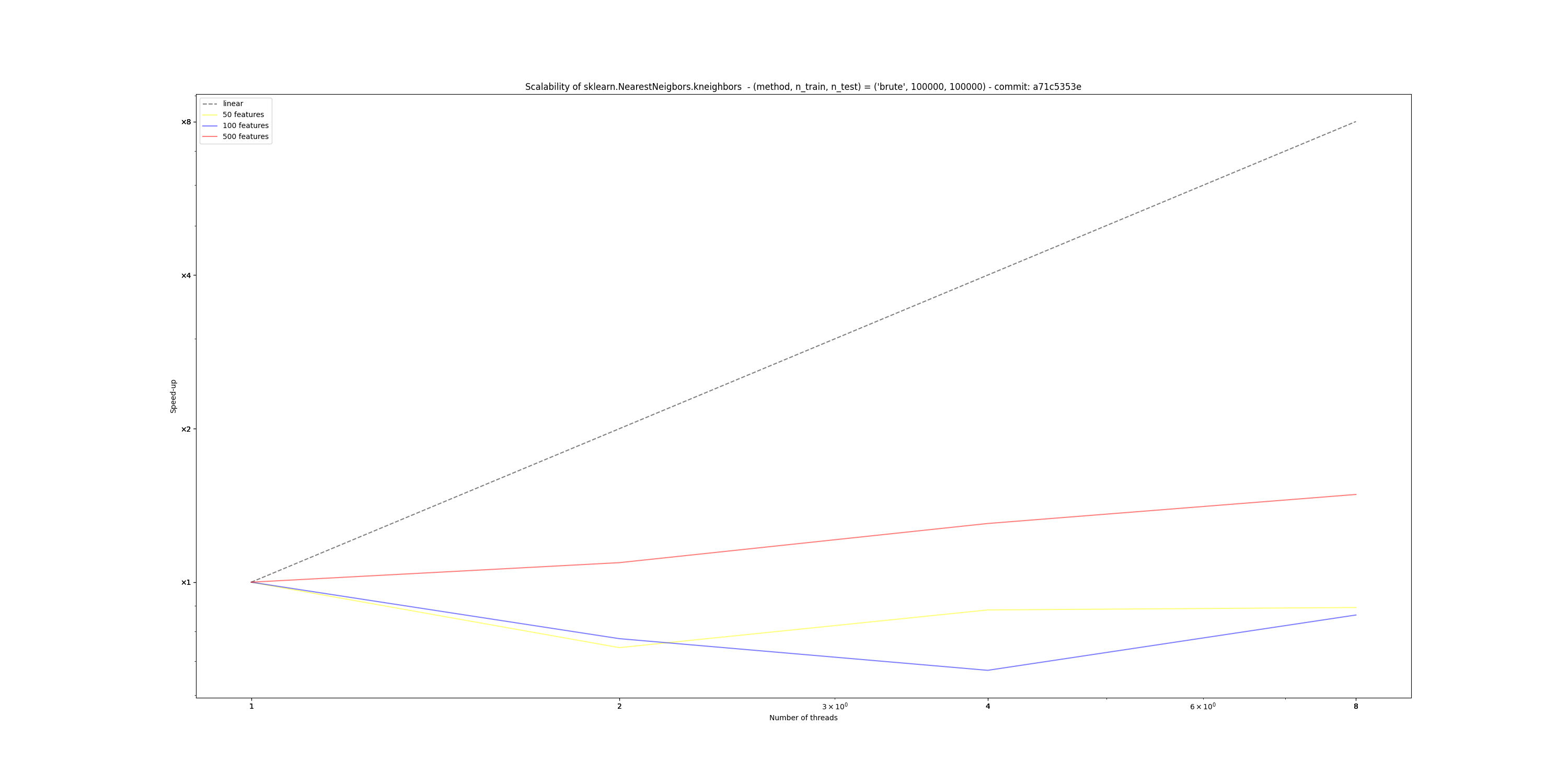
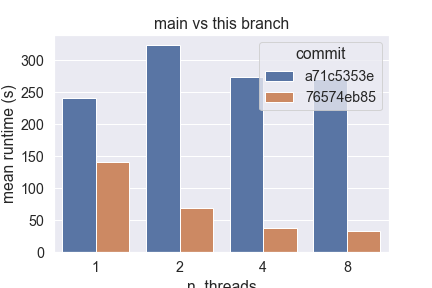
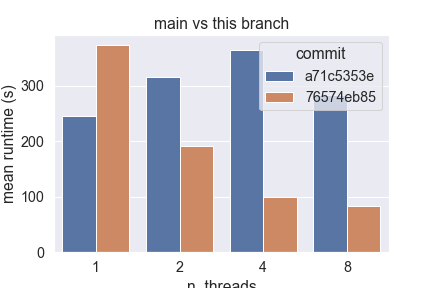
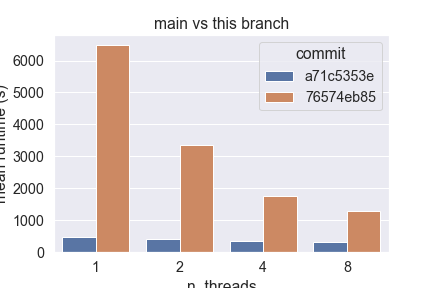
Hardware Scalability on 76574eb (using this script)The current implementation of Sparse Sparse Euclidean NearestNeighbors does not benefit from Following the design guidelines previously established, this new implementation:
This new design scales easily up to 8 cores on my laptop, unlike the current implementation that fails to do so. Below are the results obtained from running the script linked in the title, with the following parameters:
However, I also observe that the absolute amount of time taken by this implementation greatly depends on the number of features: in high dimension (> 500), this branch currently performs worse than main, so I bit more investigation is required here.
Raw data this branch
main
@Micky774 could you give me your thoughts about this PR? :) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
sklearn/metrics/_pairwise_distances_reduction/_middle_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_middle_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
| @@ -84,8 +84,7 @@ cdef class RadiusNeighbors{{name_suffix}}(BaseDistancesReduction{{name_suffix}}) | |||
| """ | |||
| if ( | |||
| metric in ("euclidean", "sqeuclidean") | |||
| and not issparse(X) | |||
| and not issparse(Y) | |||
| and not (issparse(X) ^ issparse(Y)) | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Side-note: I think it would be nice to turn in another PR is_valid_sparse_matrix, i.e.:
scikit-learn/sklearn/metrics/_pairwise_distances_reduction/_dispatcher.py
Lines 101 to 111 in 40a6afb
| def is_valid_sparse_matrix(X): | |
| return ( | |
| isspmatrix_csr(X) | |
| and | |
| # TODO: support CSR matrices without non-zeros elements | |
| X.nnz > 0 | |
| and | |
| # TODO: support CSR matrices with int64 indices and indptr | |
| # See: https://github.com/scikit-learn/scikit-learn/issues/23653 | |
| X.indices.dtype == X.indptr.dtype == np.int32 | |
| ) |
into is_supported_sparse_matrix in the global scope of base.pyx and propagated in place of scipy.sparse.{issparse,isspmatrix_csr} in sklearn.metrics._pairwise_distances_reduction.
sklearn/metrics/_pairwise_distances_reduction/_middle_term_computer.pyx.tp
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_middle_term_computer.pyx.tp
Show resolved
Hide resolved
Co-authored-by: Julien Jerphanion <git@jjerphan.xyz>
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
LGTM with pending acceptance for merge due to a duplicated allocation and cast as explained in the comment bellow.
Co-authored-by: Julien Jerphanion <git@jjerphan.xyz>
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
LGTM. Let's solve the TODOs in another PR.
Co-authored-by: Julien Jerphanion <git@jjerphan.xyz>
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
LGTM. Just a few nitpick to make the not-XOR based conditions easier to grasp as I think it's quite rare to see such constructs in our code.
sklearn/metrics/_pairwise_distances_reduction/_middle_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_middle_term_computer.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_radius_neighbors.pyx.tp
Outdated
Show resolved
Hide resolved
sklearn/metrics/_pairwise_distances_reduction/_radius_neighbors.pyx.tp
Outdated
Show resolved
Hide resolved
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
We are almost there!
|
Congrats @Vincent-Maladiere! 🎵 🎉 👏 That is significant performance improvements for many estimators! 🎉 |
Reference Issues/PRs
Relates #23585 @jjerphan
What does this implement/fix? Explain your changes.
MiddleTermComputergeneralizingGEMMTermComputerto handle the computation ofMiddleTermComputeris extended by:DenseDenseMiddleTermComputerwhen both X and Y are dense (whose implementation originates fromGEMMTermComputer)SparseSparseMiddleTermComputerwhen both X and Y are CSR. This components relies on a Cython routine.EuclideanArgKminandEuclideanRadiusNeighborsto only have them adhere toMiddleTermComputeris_usable_forinBaseDistanceReductionDispatcherto add the CSR-CSR case.computeinArgKminandRadiusNeighborsto select theEuclideanclass for the CSR-CSR case_sqeuclidean_row_normsAny other comments?
For benchmark results, see the following comments:
Euclideanspecializations ofPairwiseDistancesReductions#24556 (comment)Euclideanspecializations ofPairwiseDistancesReductions#24556 (comment)Euclideanspecializations ofPairwiseDistancesReductions#24556 (comment)