The COVID-19 Forecasting Benchmark aims to provide an evaluation platform to AI/ML researchers interested in epidemic forecasting. Submit your own forecasts and find out where you stand compared to other AI/ML based forecasts and expert COVID-19 forecasts.
See current evaluations at: https://scc-usc.github.io/covid19-forecast-bench
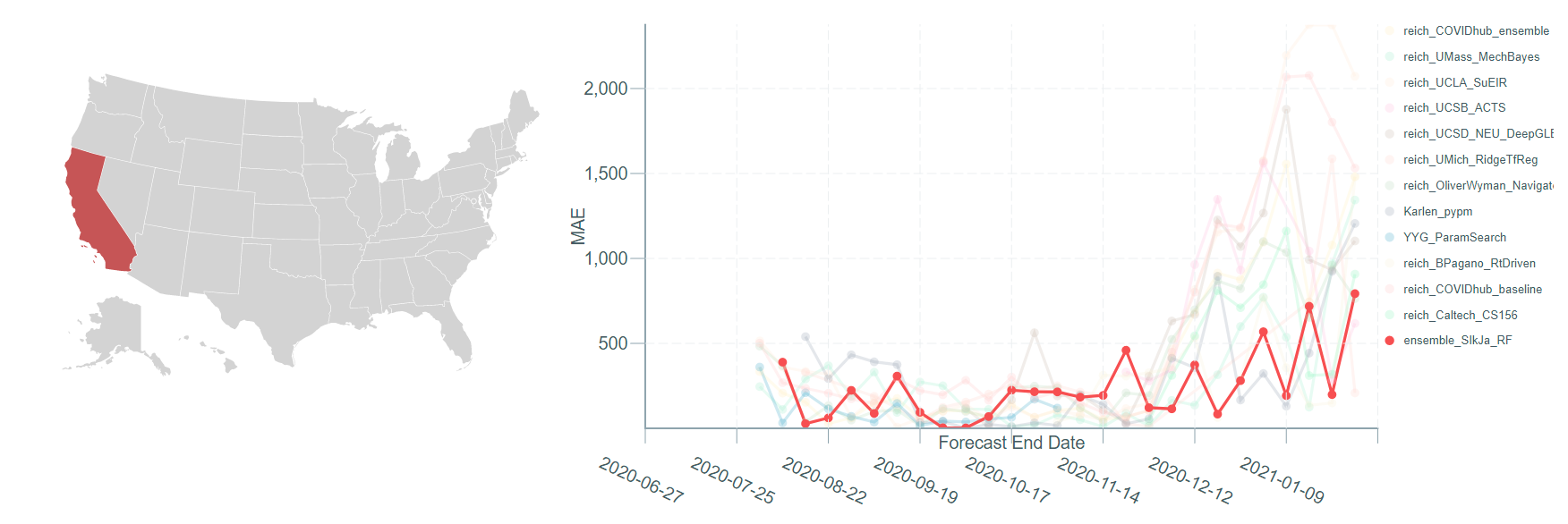
The existing comparisons during COVID-19 are those of forecasts (forecasting teams) rather than forecasting methodologies. Instead of "who did well", we are interested in "what worked well" over the duration of the pandemic. The goal of this work is to compare various methodologies and create a benchmark against which new methodologies can be evaluated. This work will also identify various decisions that affect the accuracy of forecasts from the submit methodologies.
We acknowledge that a proper evaluation is non-trivial. When designing a new methodology, bringing the entire evaluation to one error number is difficult. One has to assess the performance over time, and the difficulty of forecasting is likely to be different over the period. The benchmark, as the performances of collection of methodologies over time, can guide in a better assessment. We encourage discussions related to evaluation metrics. Please feel free to post your suggestions on Github Issues regarding evaluation metric or anything else regarding the benchmark.
We invite the interested teams to submit forecasts generated by their methodology by issuing a pull request.
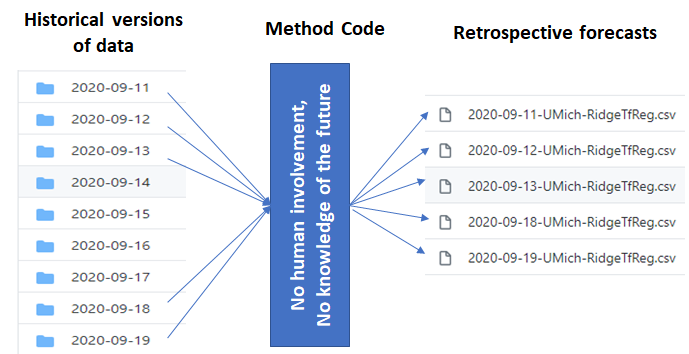
- Data to use: While generating retrospective forecasts, please ensure that you are only using data that was available before then. Our Github repo provides the version of case and death data available on previous days . The method should treat them as "separate datasets" without any knowledge of the date of the data, i.e., the method should be consistent irrespective of the date. You may use the historical versions of the JHU data available at our other repo in time-series format. Other data sources may be used as long as you can guarantee that no "foresight" was used.
- Submission format :The format of the file should be exactly like the submissions for Reich Lab's forecast hub. Please follow the naming convention: [Date of forecast (YYYY-MM-DD)]-[Method_Name].csv.
- Forecast dates : We will take the retrospective forecasts for any range of time, starting in July until the present.
- Forecast locations : Currently, we are only accepting case and death forecasts for US state, county, and national-level and national-level for other countries. More locations will be addressed in the future.
- Forecast horizon: The forecasts are expected to be incident cases forecasts per week observed on a Sunday for 1, 2, 3, and 4-week ahead. One week ahead forecast generated after a Monday is to be treated as the Sunday after the next one. This is in accordance with the Reich Lab's forecasting hub.
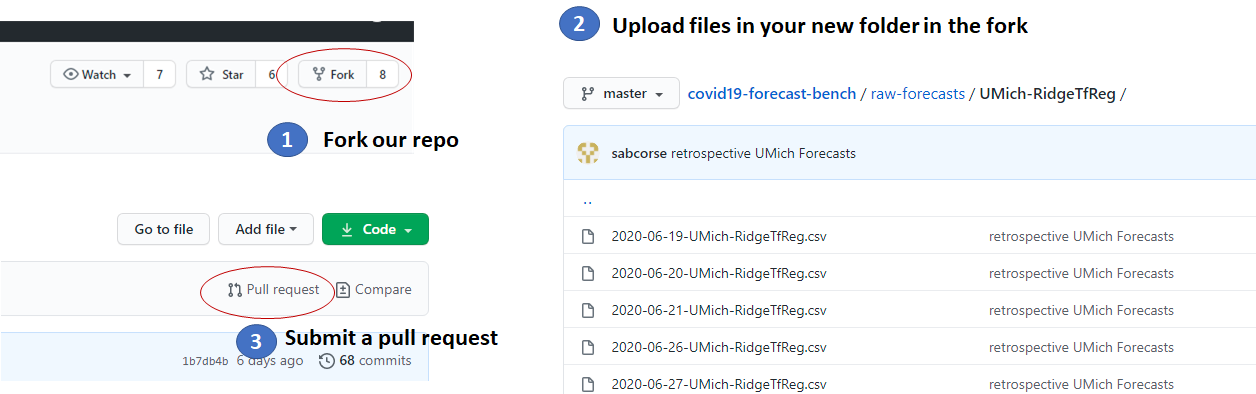
- Where to upload files : Please add your files in the folder "raw-forecasts/" in your fork of our repo and submit a pull request. It can be done directly using your browser, cloning the repo is not needed:
- Methodology Description: In a file named metadata-[Method Name].csv, please provide a short description of your approach, mentioning at least the following:
- Modeling technique: Generative (SIR, SEIR, ...), Discriminative (Neural Networks, ARIMA, ...), ...
- Learning approach: Bayesian, Regression (Quadratic Programming, Gradient Descent, ...), ...
- Pre-processing: Smoothing (1 week, 2, week, auto), anomaly detection, ...
- Submitting forecasts from multiple methodologies: If you are submitting forecasts for multiple methodolgies, please ensure there is something in their metadata descriptions that differentiates them. Please note that any change in your approach, including data pre-processing and hand-tuning a parameter counts as a different methodology. You can alter your method name to mark the distinction such as by appending an appropriate suffix.
- License: We encourage you to make your submission under the opensource MIT license.
We are grateful for your support and look forward to your contributions!
Ajitesh Srivastava (ajiteshs[AT]usc[DOT]]edu)