SCTransform for Python - interfaces with Scanpy
See demo.
We recommend using conda for installing pySCTransform.
conda create -n pysct louvain scanpy
conda activate pysct
pip install git+https://github.com/saketkc/pysctransform.git@glmgpIf you would like to use glmGamPoi, a faster estimator, rpy2 and glmGamPoi need to be installed as well:
conda create -n pysct louvain scanpy rpy2 bioconductor-glmgampoi
conda activate pysct
pip install git+https://github.com/saketkc/pysctransform.gitimport scanpy as sc
from pysctransform import SCTransform
pbmc3k = sc.read_h5ad("./pbmc3k.h5ad")
# Get pearson residuals for 3K highly variable genes
residuals = SCTransform(pbmc3k, var_features_n=3000)
pbmc3k.obsm["pearson_residuals"] = residuals
# Peform PCA on pearson residuals
pbmc3k.obsm["X_pca"] = sc.pp.pca(pbmc3k.obsm["pearson_residuals"])
# Clustering and visualization
sc.pp.neighbors(pbmc3k, use_rep="X_pca")
sc.tl.umap(pbmc3k, min_dist=0.3)
sc.tl.louvain(pbmc3k)
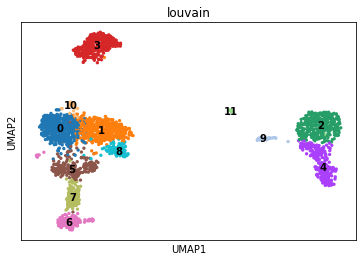
sc.pl.umap(pbmc3k, color=["louvain"], legend_loc="on data", show=True)# Perform variance stabilization using 'v2' regularization
from pysctransform import vst
from pysctransform.plotting import plot_residual_var
vst_out_3k = vst(umi = pbmc3k.X.T,
gene_names=pbmc3k.var_names.tolist(),
cell_names=pbmc3k.obs_names.tolist(),
method="fix-slope",
exclude_poisson=True
)
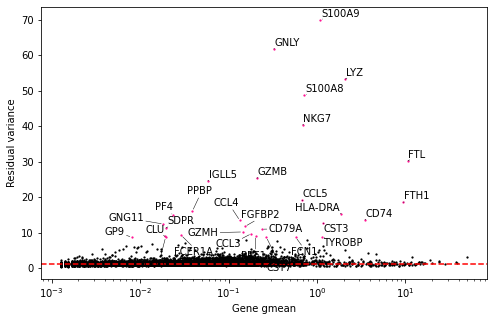
plot_residual_var(vst_out_3k)batch_varis currently not supported