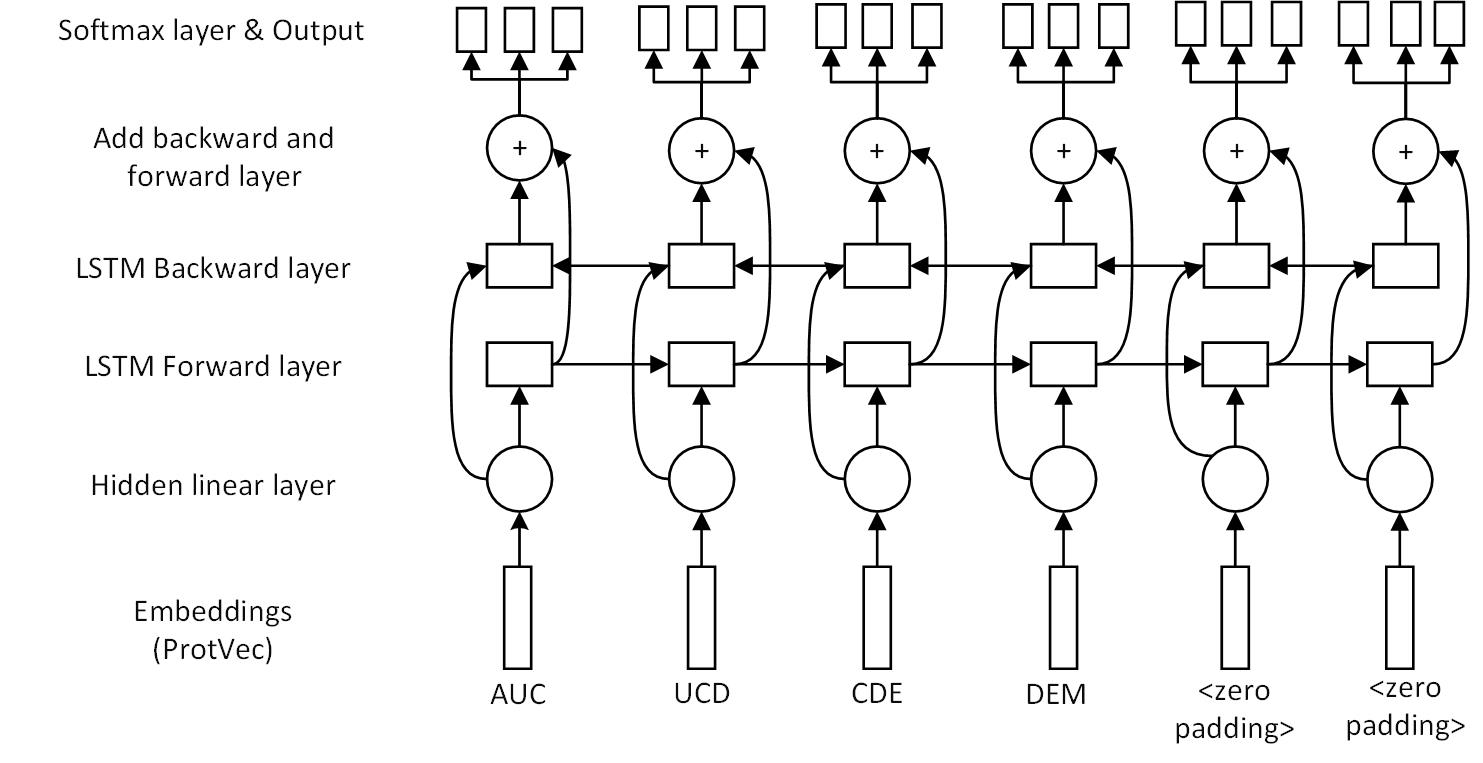
Multitask learning (secondary structure prediction, b-values prediction, solvent-accessibility prediction) can improve the prediction accuracy of protein secondary structure.
- We have to face with the class imbalance problem
- "foldername_cv": 5 fold cross validation
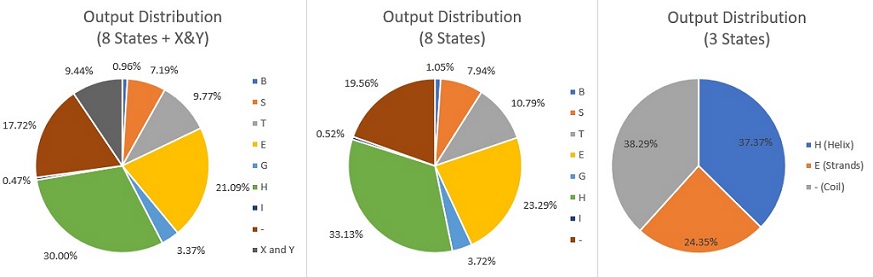
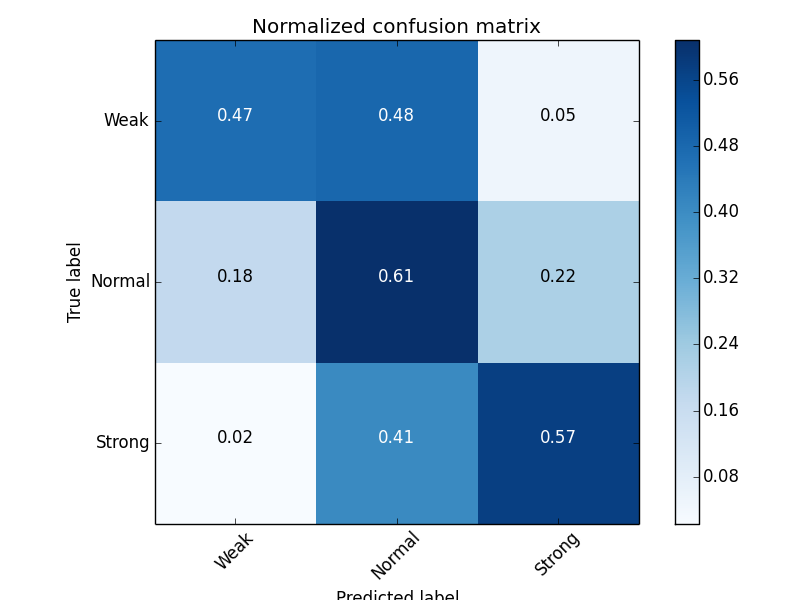
- Distribution of outputs:
The copyright belongs to http://rostlab.org/. It can not be public.
Using Protvec (3-gram) and follow the vector addition rule. For example:
TNCDE = UTN + TNC + NCD + CDE + DEU
-
Secondary Structure accuracy (3 states): 69.0%
-
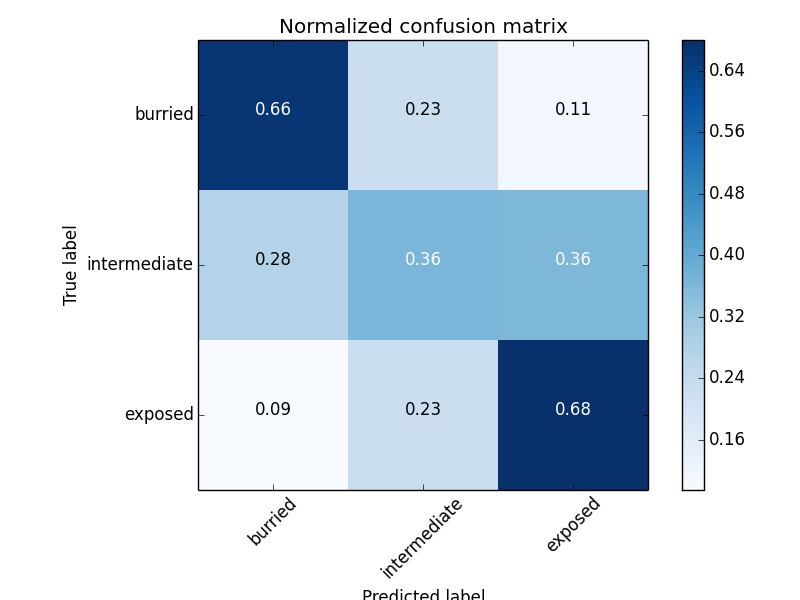
Solvent Accessibility accuracy (3 states): 54.6%
-
B-values accuracy (3 states): 59.1%
-
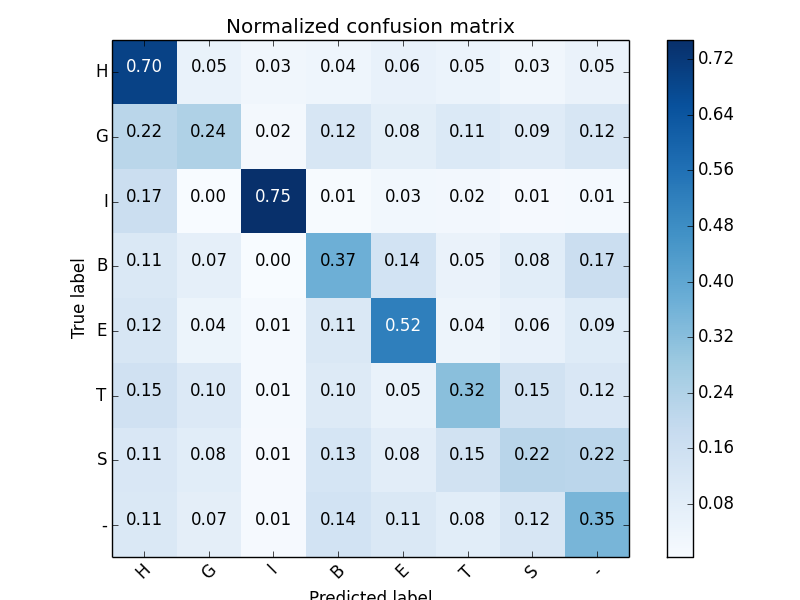
Secondary Structure accuracy (8 states): 0.476
-
Solvent Accessibility accuracy (3 states): 0.548
-
B-values accuracy (3 states): 0.598
- Secondary structure
- Solvent accessibility
- b-values
- python 2.7
- tensorflow 1.4.0
- ProtVec
Go into each subfolder and run the code following:
- python lstm.py
Binh Do
This project is licensed under the MIT License