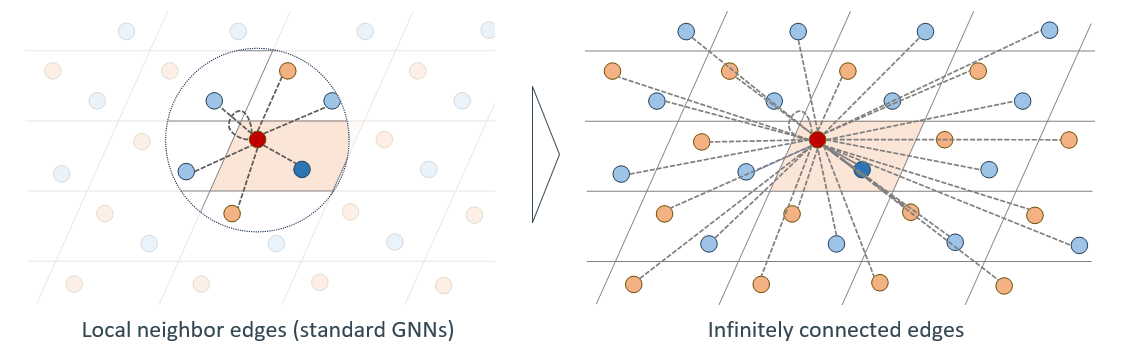
Crystalformer: Infinitely Connected Attention for Periodic Structure Encoding
Tatsunori Taniai, Ryo Igarashi, Yuta Suzuki, Naoya Chiba, Kotaro Saito, Yoshitaka Ushiku, and Kanta Ono
In The Twelfth International Conference on Learning Representations (ICLR 2024)
@inproceedings{taniai2024crystalformer,
title = {Crystalformer: Infinitely Connected Attention for Periodic Structure Encoding},
author = {Tatsunori Taniai and
Ryo Igarashi and
Yuta Suzuki and
Naoya Chiba and
Kotaro Saito and
Yoshitaka Ushiku and
Kanta Ono
},
booktitle = {The Twelfth International Conference on Learning Representations},
year = {2024},
url = {https://openreview.net/forum?id=fxQiecl9HB}
}
cd docker/pytorch21_cuda121
docker build -t main/crystalformer:latest .
docker run --gpus=all --name crystalformer --shm-size=2g -v ../../:/workspace -it main/crystalformer:latest /bin/bashIn the docker container:
cd /workspace/data
python download_megnet_elastic.py
python downlad_jarvis.py # This step may take 1 hour and can be skipped for simple testing.Download pretrained weights: [GoogleDrive] In the `/workspace' directory in the docker container:
unzip weights.zip
. demo.shCurrently, pretrained models for MEGNET's bandgap and e_form with 4 or 7 attention blocks are available.
In the `/workspace' directory in the docker container:
CUDA_VISIBLE_DEVICES=0 python train.py -p latticeformer/default.json \
--save_path result \
--n_epochs 500 \
--experiment_name demo \
--num_layers 4 \
--value_pe_dist_real 64 \
--target_set jarvis__megnet-shear \
--targets shear \
--batch_size 128 \
--lr 0.0005 \
--model_dim 128 \
--embedding_dim 128 \
Setting --value_pe_dist_real 0 yields the "simplified model" in the paper.
In the `/workspace' directory in the docker container:
CUDA_VISIBLE_DEVICES=0,1 python train.py -p latticeformer/default.json \
--save_path result \
--n_epochs 500 \
--experiment_name demo \
--num_layers 4 \
--value_pe_dist_real 64 \
--target_set jarvis__megnet-shear \
--targets shear \
--batch_size 128 \
--lr 0.0005 \
--model_dim 128 \
--embedding_dim 128 \
Currently, the throughput gain by multi-gpu training is limited. Suggest 2 or 4 GPUs at most.
| target_set | targets | Unit | train | val | test |
|---|---|---|---|---|---|
| jarvis__megnet | e_form | eV/atom | 60000 | 5000 | 4239 |
| jarvis__megnet | bandgap | eV | 60000 | 5000 | 4239 |
| jarvis__megnet-bulk | bulk | log(GPA) | 4664 | 393 | 393 |
| jarvis__megnet-shear | shear | log(GPA) | 4664 | 392 | 393 |
| dft_3d_2021 | formation_energy | eV/atom | 44578 | 5572 | 5572 |
| dft_3d_2021 | total_energy | eV/atom | 44578 | 5572 | 5572 |
| dft_3d_2021 | opt_bandgap | eV | 44578 | 5572 | 5572 |
| dft_3d_2021-mbj_bandgap | mbj_bandgap | eV | 14537 | 1817 | 1817 |
| dft_3d_2021-ehull | ehull | eV | 44296 | 5537 | 5537 |
Use the following hyperparameters:
- For the
jarvis__megnetdatasets:--n_epochs 500 --batch_size 128 - For the
dft_3d_2021-mbj_bandgapdataset:--n_epochs 1600 --batch_size 256 - For the other
dft_3d_2021datasets:--n_epochs 800 --batch_size 256
General training hyperparameters:
n_epochs(int): The number of training epochs.batch_size(int): The batch size (i.e., the number of materials per training step).loss_func(L1|MSE|Smooth_L1): The regression loss function form.optimizer(adamw|adam|): The choice of optimizer.adam_betas(floats): beta1 and beta2 of Adam and AdamW.lr(float): The initial learning rate. The default setting (5e-4) works mostly the best.lr_sch(inverse_sqrt_nowarmup|const): The learning rate schedule.inverse_sqrt_nowarmupsets learning rate tolr*sqrt(t/(t+T))where T is specified bysch_params.constuses a constant learning ratelr.
Final MLP's hyperparameters:
embedding_dim(ints): The intermediate dims of the final MLP after pooling, defining Pooling-Repeat[Linear-ReLU]-FinalLinear. The default setting (128) defines Pooling-Linear(128)-ReLU-FinalLinear(1).norm_type(no|bn): Whether or not use BatchNorm in MLP.
Transformer's hyperparameters:
num_layers(int): The number of self-attention blocks. Should be 4 or higher.model_dim(int): The feature dimension of Transformer.ff_dim(int): The intermediate feature dimension of the feed-forward networks in Transformer.head_num(int): The number of heads of multi-head attention (HMA).
Crystalformer's hyperparameters.
scale_real(float or floats): "r_0" in the paper. (Passing multiple values allows different settings for individual attention blocks.)gauss_lb_real(float): The bound "b" for the rho function in the paper.value_pe_dist_real(int): The number of radial basis functions (i.e., edge feature dim "K" in the paper). Should be a multiple of 16.value_pe_dist_max(float): "r_max" in the paper. A positive value directly specifies r_max in Å, while a negative value specifies r_max via r_max = (-value_pe_dist_max)*scale_real.domain(real|multihead|real-reci): Whether use reciprocal-space attention by parallel MHA (multihead) or block-wisely interleaving between real and reciprocal space (real-reci). When reciprocal-space attention is used,scale_reciandgauss_lb_recican also be specified.
For each of train, val, and test splits, make a list of dicts containing pymatgen's Structures and label values:
- list
- dict
- 'structure': pymatgen.core.structure.Structure
- 'property1': a float value of
propety1of this structure - 'property2': a float value of
propety2of this structure - ...
- dict
Dump the list of each split in a directory with your dataset name as
import os
import pickle
target_set = 'your_dataset_name'
split = 'train' # or 'val' or 'test'
os.makedirs(f'data/{target_set}/{split}/raw', exist_ok=True)
with open(f'data/{target_set}/{split}/raw/raw_data.pkl', mode="wb") as fp:
pickle.dump(data_list, fp)Then, you can specify your dataset and its target property name as
python train -p latticeformer/default.json \
--target_set your_dataset_name \
--targets [property1|property2] \