The CellOrganizer project provides tools for
- learning generative models of cell organization directly from images
- storing and retrieving those models
- synthesizing cell images (or other representations) from one or more models
Model learning captures variation among cells in a collection of images. Images used for model learning and instances synthesized from models can be two- or three-dimensional static images or movies.
CellOrganizer can learn models of
- cell shape
- nuclear shape
- chromatin texture
- vesicular organelle size, shape and position
- microtubule distribution.
These models can be conditional upon each other. For example, for a given synthesized cell instance, organelle position is dependent upon the cell and nuclear shape of that instance.
Cell types for which generative models for at least some organelles have been built include human HeLa cells, mouse NIH 3T3 cells, and Arabidopsis protoplasts. Planned projects include mouse T lymphocytes and rat PC12 cells.
- Created method slml2slml. The method joins together multiple model files.
- Updated methods for CellOrganizer for Galaxy.
- Fixed several small issues that accelerate synthesis.
- Fixed issues with the creation of OME.TIFFs with ROIs.
- Created method slml2info to replace model2info. The method creates a report with useful information from a model file. Method writes report with Matlab Publisher. When method is deployed, then it generates a report in Markdown.
- Created method slml2report. The method creates a report comparing two model files. Method writes report with Matlab Publisher. When method is deployed, then it generates a report in Markdown.
- Created demos demo2D08 and demo2D09 to show how to make a report from a PCA framework model.
- Fixed problem with demo3D46. Unable to synthesize image from CSGO model when combined with a vesicle model.
- Fixed problem with demo3D35. Unable to produce figure.
- Included new model class/type: constructive_geometry/half-ellipsoid
- Included new model class/type: framework/pca
- Included support for OME.TIFF with regions of interest
- Included bash scripts to run demos from compiled versions of the main functions
- img2slml now checks the combination of model class and type for every submodel before attempting to extract parameters from image
- Improved pipeline so that CellOrganizer will stop computation and report to user if no images are found in the path or if software fails to extract parameters.
- Added demo3D44 to show how to synthesize from a model class/type constructive_geomertry/half-ellipsoid
- Added a battery of unit test for demos using Matlab testing framework
- Added demo3D45 to show how to use OME.TIFF files with ROIs.
- Added demo2D05, demo2D06, demo2D07 to show how to train and synthesize from a classtype framework/pca model
The following demo scripts are included in the image.
| Demo Name | Training | Synthesis |
|---|---|---|
| demo2D00 | X | |
| demo2D01 | X | |
| demo2D02 | X | |
| demo2D04 | X | |
| demo2D05 | X | |
| demo3D00 | X | |
| demo3D01 | X | |
| demo3D03 | X | |
| demo3D04 | X | |
| demo3D06 | X | |
| demo3D07 | X | |
| demo3D08 | X | |
| demo3D09 | X | |
| demo3D11 | X | |
| demo3D12 | X | |
| demo3D19 | ||
| demo3D20 | X | |
| demo3D47 |
The demos in the table above are the same demos included in the Matlab distribution.
Installing Docker is beyond the scope of this document. To learn about Docker Community Edition (CE), click here.
The easiest way to download an image and run a container is to use [Kitematic(https://kitematic.com/).
- To install Kitematic, click here.
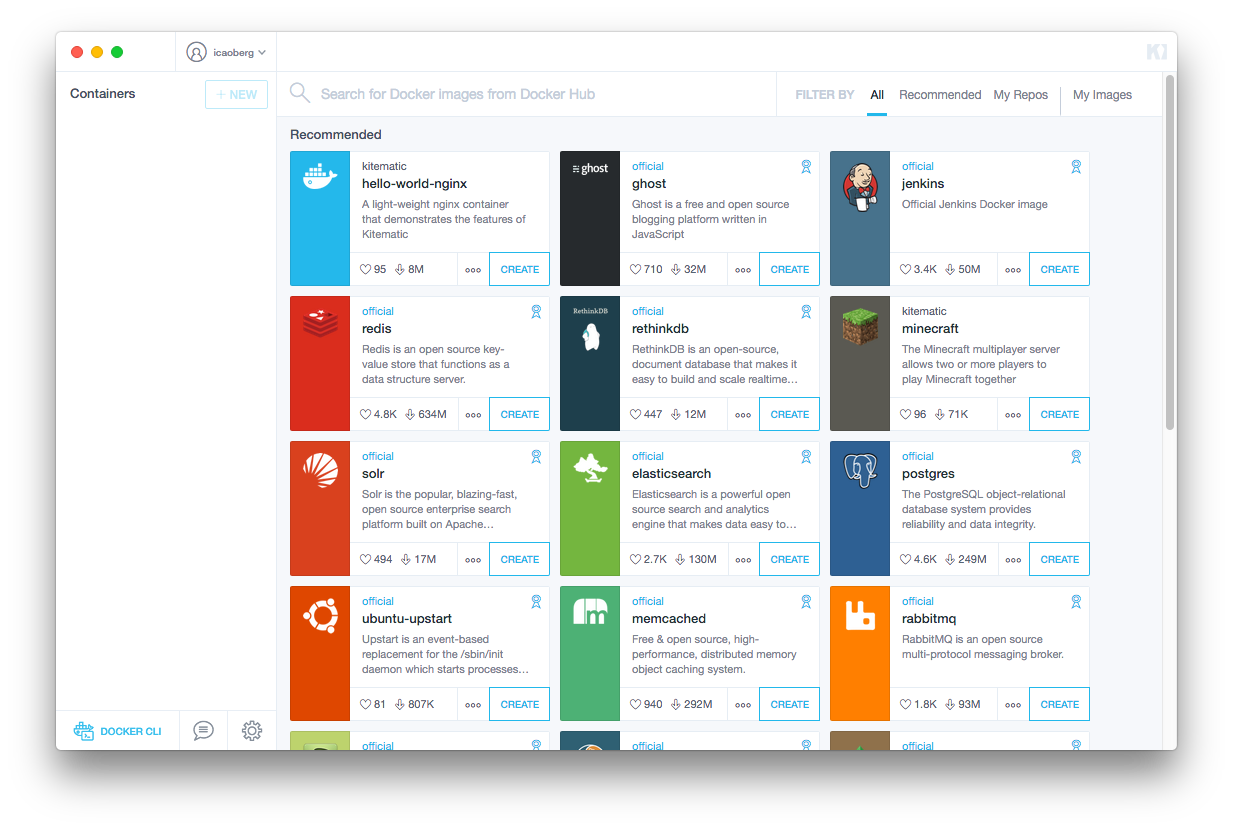
Running Kitematic will open a window that looks like this
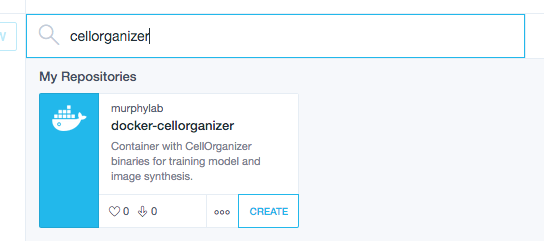
Use the searchbar to search for cellorganizer
and click Create.
To build an image using the Dockerfile in this repository, run the command
➜ docker build -t "murphylab/cellorganizer" .
The previous step should build an image
➜ docker container ls -a
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
48dde52f2bc8 murphylab/cellorganizer "/bin/bash -c 'pyt..." 45 seconds ago Exited (0) 39 seconds ago frosty_wescoff
To run a container using the image above
➜ docker run -i -t murphylab/cellorganizer
The container comes with
- CellOrganizer binaries
- Generative models
- Murphy Lab 2D/3D HeLa datasets
- BioFormats tools
- vim-as-an-IDE
When contributing to this repository, please first discuss the change you wish to make via issue, email, or any other method with the owners of this repository before making a change.
Support for CellOrganizer has been provided by grants GM075205, GM090033 and GM103712 from the National Institute of General Medical Sciences, grants MCB1121919 and MCB1121793 from the U.S. National Science Foundation, by a Forschungspreis from the Alexander von Humboldt Foundation, and by the Freiburg Institute for Advanced Studies.
Copyright (c) 2007-2019 by the Murphy Lab at the Computational Biology Department in Carnegie Mellon University