This repository hosts the code and guides for the pipelines used in the paper A deep learning-based toolkit for 3D nuclei segmentation and quantitative analysis in cellular and tissue context. It is structured in to four folders:
- stardist/ contains a 3D StarDist training and inference pipeline,
run-stardist. - plantseg/ contains configuration files for training and inference with PlantSeg.
- cellpose/ contains scripts for training and inference with Cellpose.
- evaluation/ contains modules for evaluating the segmentation results.
See run-stardist's README.md for more details.
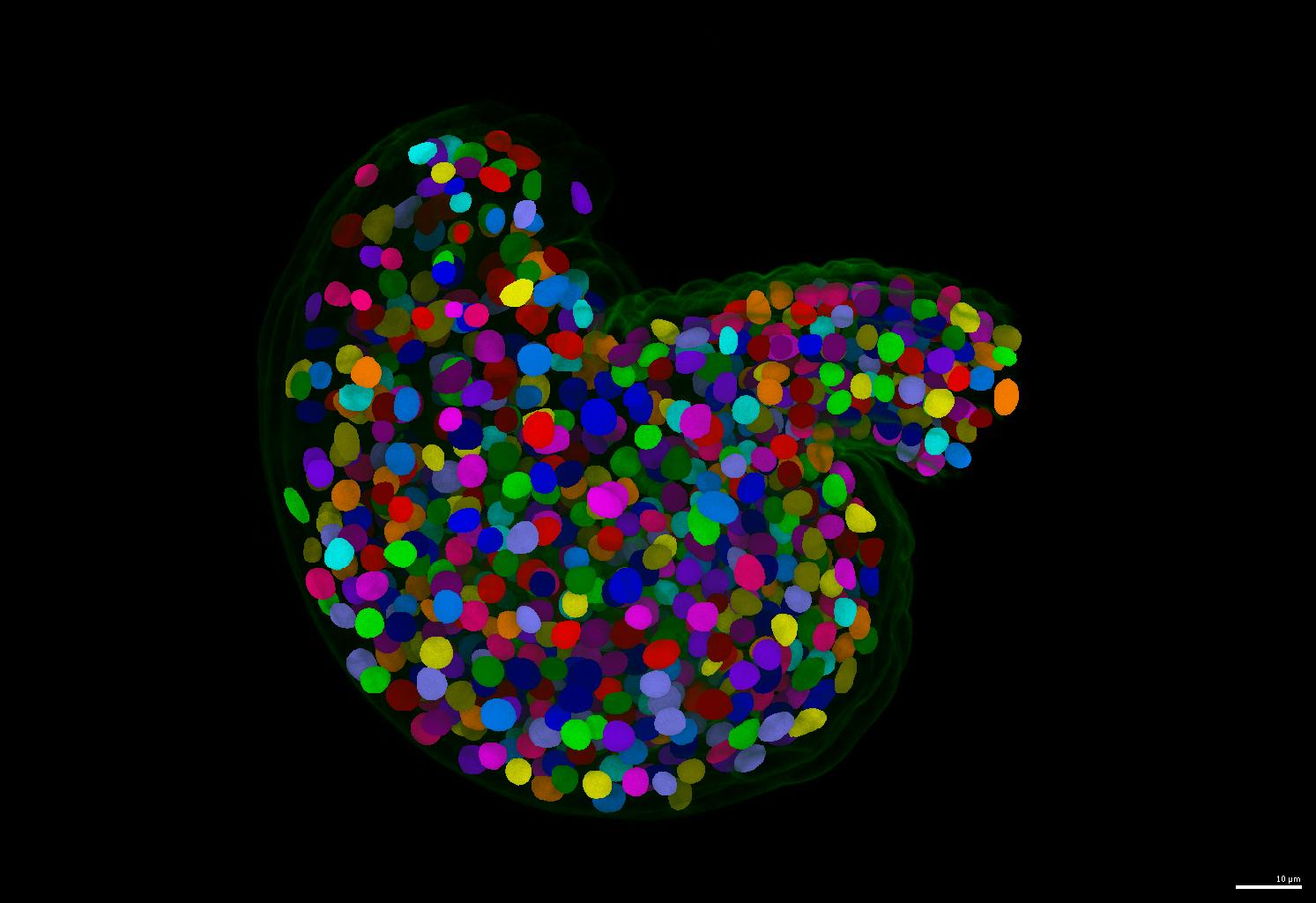
This is one of the most important contribution of this repository. If your nuclei are more or less uniform in shape, please consider using the run-stardist pipeline in this repository. It generate separate and round instance segmentation masks for your nuclei images.
- The code and tutorial for running StarDist inference is in the
stardist/folder - The pretrained model is automatically downloaded during inference (also available at BioImage.IO: StarDist Plant Nuclei 3D ResNet)
- An example of segmentation results is shown below.
See PlantSeg's README.md for more details.
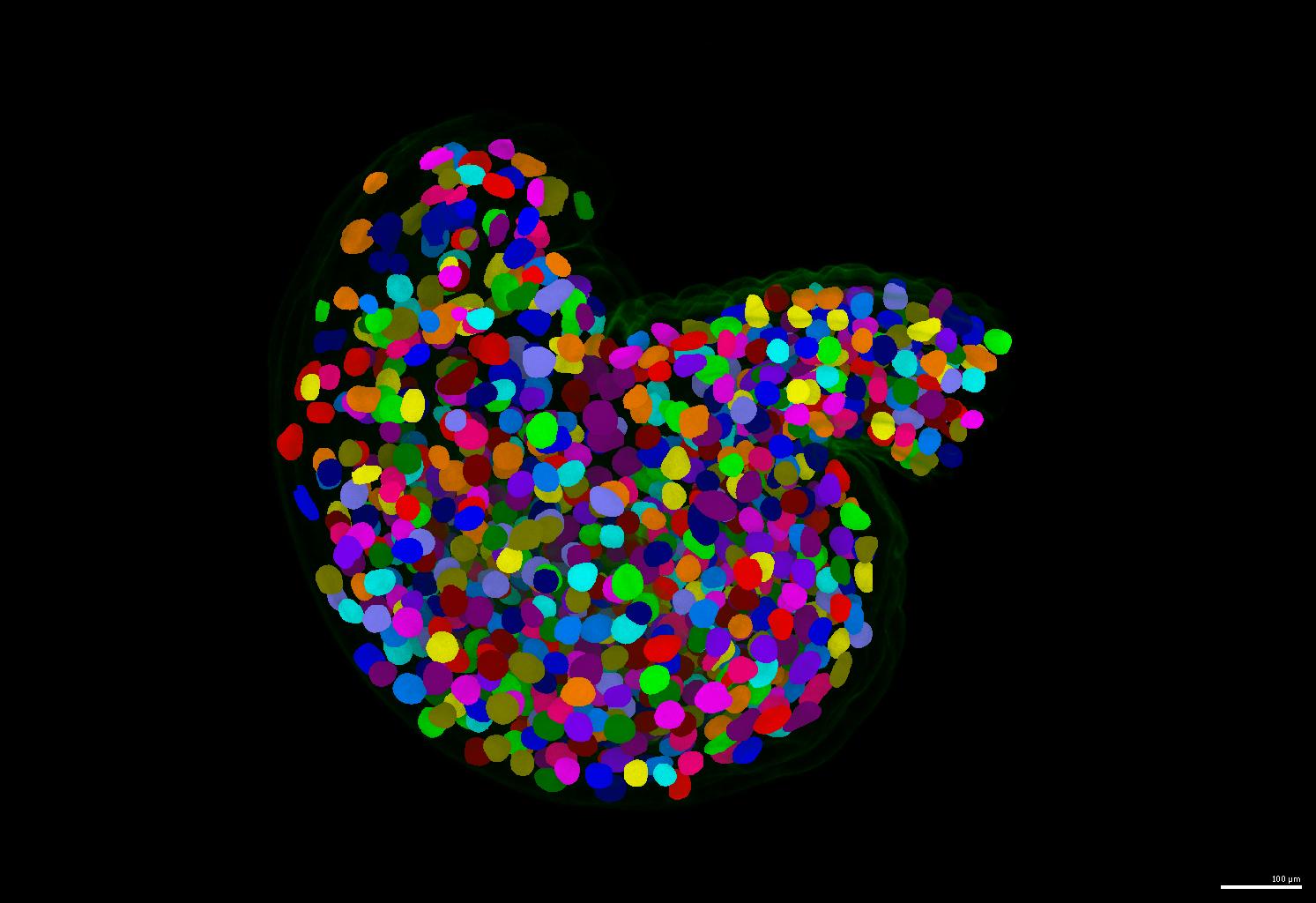
If your nuclei have irregular shapes, please consider using the PlantSeg pipeline. It generates instance masks for your nuclei images regardless of their nucleus size and shape.
- The code and tutorial for running PlantSeg inference is in the
plantseg/folder - The pretrained model is automatically downloaded during inference (also available at BioImage.IO: PlantSeg Plant Nuclei 3D UNet)
- An example of segmentation results is shown below.
See Cellpose's README.md for more details.
- The guide for running Cellpose inference and training is in the
cellpose/folder
The training data is publicly available on Zenodo at [TODO](to be published after paper submission).
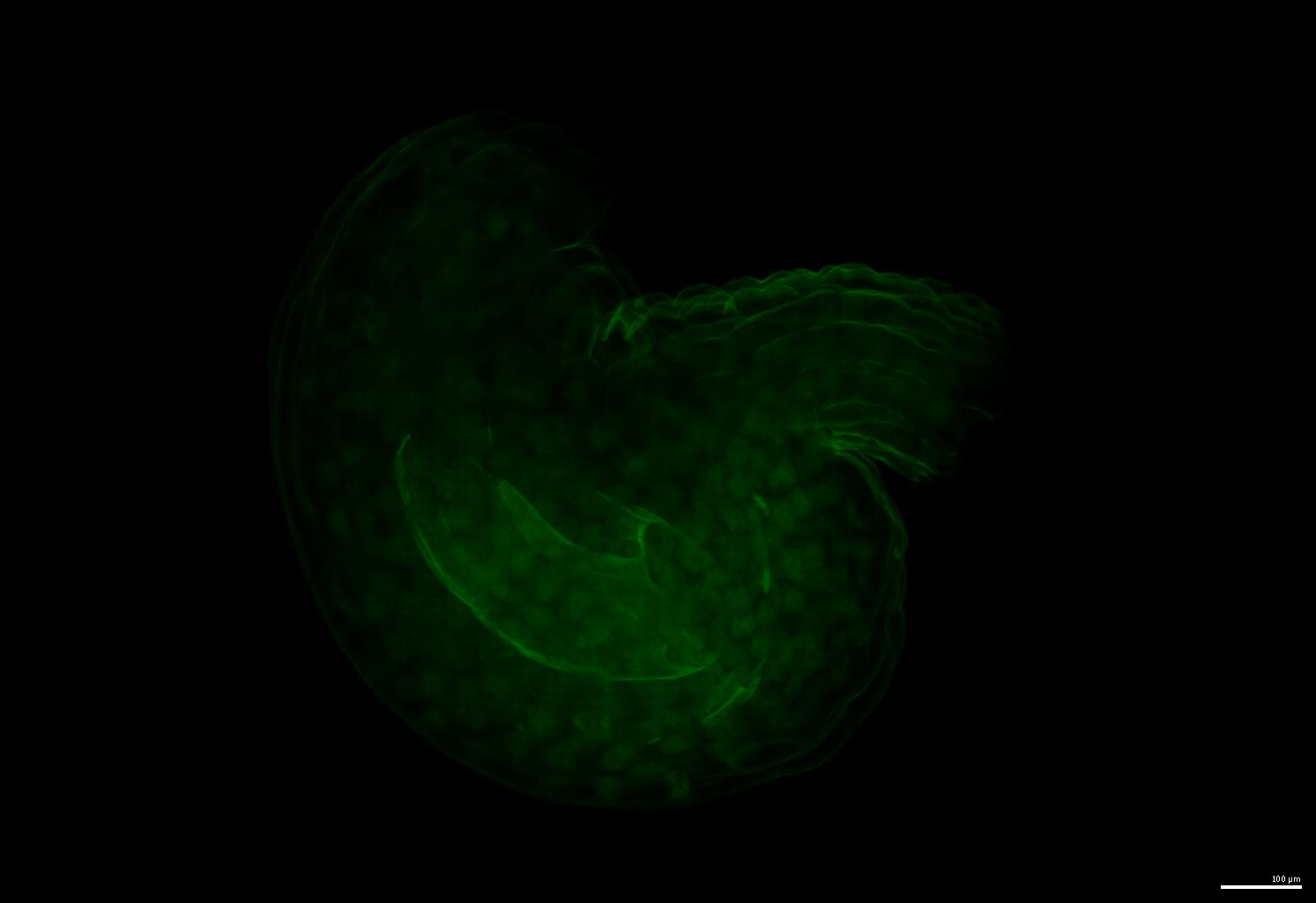
An example of the raw image:
Some key information about the training data is listed below:
original_voxel_size = { # z, y, x
1135: [0.28371836501901143, 0.12678642066720086, 0.12678642066720086], # validation
1136: [0.2837183895131086, 0.12756971653115998, 0.12756971653115998], # training
1137: [0.2837183895131086, 0.1266211463645486, 0.1266211463645486 ], # training
1139: [0.2799036917562724, 0.12674335484590543, 0.12674335484590543], # training
1170: [0.27799632231404964, 0.12698523961670266, 0.12698522349145364], # training
} # [0.2837, 0.1268, 0.1268] is taken as the median
original_median_extents = { # z, y, x
1135: [16, 32, 33], # validation
1136: [16, 32, 32], # training
1137: [16, 32, 32], # training
1139: [16, 32, 33], # training
1170: [16, 29, 30], # training
'average':
} # [16, 32, 32] is taken as the medianNote for training Cellpose: The best image form for training StarDist and PlantSeg models are the original forms, i.e. the linked dataset is the one that provide the best results. However, to train Cellpose which only takes 2D training data, the images are prepared to be 2D slices of the rescaled isotropic 3D images. The 2D slices includes all XY, XZ and YZ slices ordered randomly by a random prefix in the file name. The 2D slices are saved as TIFF files.
Both HDF5 files and TIFF files can be directly used for both run-stardist and plant-seg inference. Go to the respective folders's README.md for more details.
If you find this work useful, please cite our paper and the respective tools' papers:
@article {Vijayan2024.02.19.580954,
author = {Athul Vijayan and Tejasvinee Atul Mody and Qin Yu and Adrian Wolny and Lorenzo Cerrone and Soeren Strauss and Miltos Tsiantis and Richard S. Smith and Fred Hamprecht and Anna Kreshuk and Kay Schneitz},
title = {A deep learning-based toolkit for 3D nuclei segmentation and quantitative analysis in cellular and tissue context},
elocation-id = {2024.02.19.580954},
year = {2024},
doi = {10.1101/2024.02.19.580954},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/02/21/2024.02.19.580954},
eprint = {https://www.biorxiv.org/content/early/2024/02/21/2024.02.19.580954.full.pdf},
journal = {bioRxiv}
}