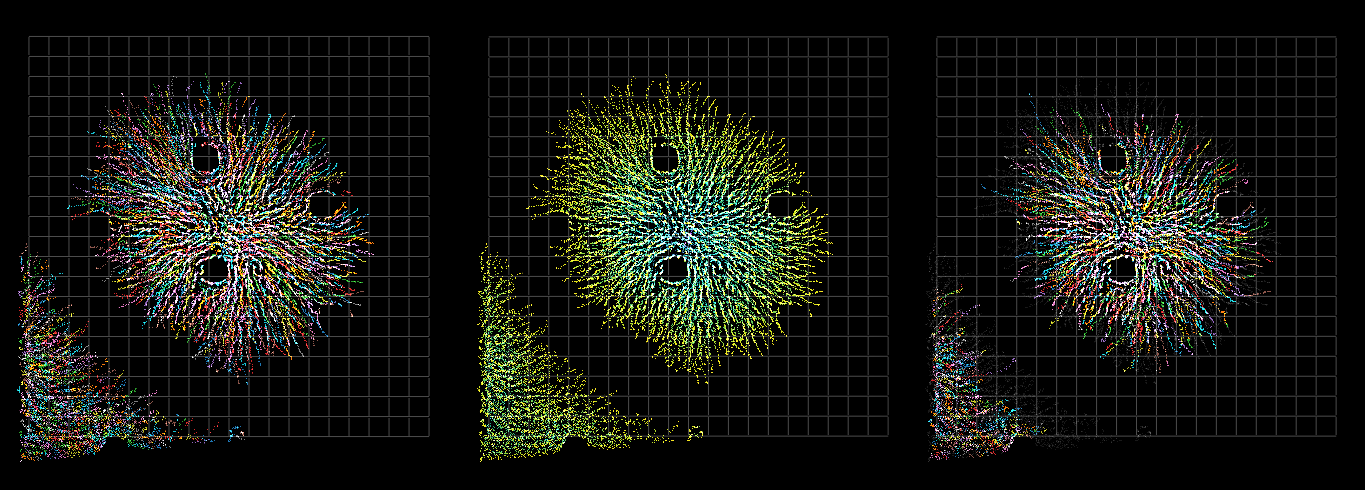
A biomovie is a temporal series of digital microscopy images that is recorded for one selected visual field in a bacterial time course experiment. We present SeeVIS, a data driven (S)egmentation-fr(EE) and automatic pipeline of methods to (VIS)ualise the growth patterns of a cell population conveyed in a biomovie. It consists of three steps 1. signal enhancement to help adjust the signal-to-noise ratio, 2. feature detection so as to draw out qualitative information about the colony development, and 3. the visualization of the colony in a feature space as a 3D space-time cube supported with three novel and appropriate color mappings. They are based on two colour palettes (Tableau10 and Viridis), are titled and functionally described as follows:
- Nominal Mapping (NM) highlights single feature trajectories (
Tableau10) - Time Mapping (TM) visually promotes the extent of the population growth over time (
Viridis) - Progeny Mapping (PM) supports the process of tracing back features to their parents (
Tableau10).
SeeVIS colour mappings demonstrated for dataset D1. Screen captures of the 3D visualization is displayed with an azimuth = 0°, an elevation = 90°, and a grid mesh for the three colour mappings: NM (left), TM (middle), and PM (right), respectively.
- German-Canadian DFG International Research Training Group GRK 1906/1
- Phenotypic Heterogeneity and Sociobiology of Bacterial Populations, DFG SPP1617.
The employed datasets are available under The Open Data Commons Attribution License (ODC-By) v1.0.
The output CSV for dataset D1 is provided: data.csv. It corresponds to the output of the first two steps for registered dataset D1 (Schlueter et al. 2015). The CSV file can be supplied to SeeVIS (see Usage).
Schlueter, J. - P., McIntosh, M., Hattab, G., Nattkemper, T. W., and Becker, A. (2015). Phase Contrast and Fluorescence Bacterial Time-Lapse Microscopy Image Data. Bielefeld University. doi:10.4119/unibi/2777409.
For better reproducibility the versions that were used for development are reported in the setup.py file and can be installed via:
$ python3 setup.py install# Set file permissions
$ chmod +x seevis.py
# Run SeeVIS on a folder containing all image files
# Formatted by channel : red, green, blue as c2, c3, c4 respectively for every time point
$ python3 seevis.py -i img_directory/
# Or on a CSV file containing feature positions
$ ./seevis.py -f data.csv -s 2
# -h, --help show this help message and exit
# -v, --version show program's version number and exit
# -i, --input run SeeVIS on the supplied directory
# -f, --file run the Visualization of SeeVIS
# -s run scheme (or colour mapping) ranging from 1 to 3 (default is 1)The visualization displays a particle in the form of a spot that scales with the view (see functions.py, line 111 and lines 393--396). The spot size can be changed according to user preferences or image modalities. The spot size has been chosen such that the particle trajectory does not contain scattered spots and such that neighboring spots do not lead to visual occlusion.
The preprocessing parameters as well as the particle paradigm are detailed in Hattab G, Wiesmann V, Becker A, Munzner T, Nattkemper TW. A novel Methodology for characterizing cell subpopulations in automated Time-lapse Microscopy. Frontiers in bioengineering and biotechnology. 2018. doi:10.3389/fbioe.2018.00017.
If you use this software, please consider citing the following paper: Hattab, G. and Nattkemper, T.W., 2018. SeeVis—3D space-time cube rendering for visualization of microfluidics image data. Bioinformatics.
@Article{hattab18seevis,
Author="Hattab, G. and Nattkemper, T. W. ",
Title="{{S}ee{V}is - 3{D} space-time cube rendering for visualization of microfluidics image data}",
Journal="Bioinformatics",
Year="2018",
Month="Oct"
}
The MIT License (MIT)
Copyright (c) Georges Hattab
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.