This repository contains the codes created or used for the analysis in the paper "Cognitive and White-Matter Compartment Models Reveal Selective Relations between Corticospinal Tract Microstructure and Simple Reaction Time"
DOI: https://doi.org/10.1523/JNEUROSCI.2954-18.2019
For more information on the codes, please contact: esin.karahan@gmail.com
web: https://ccbrain.org/
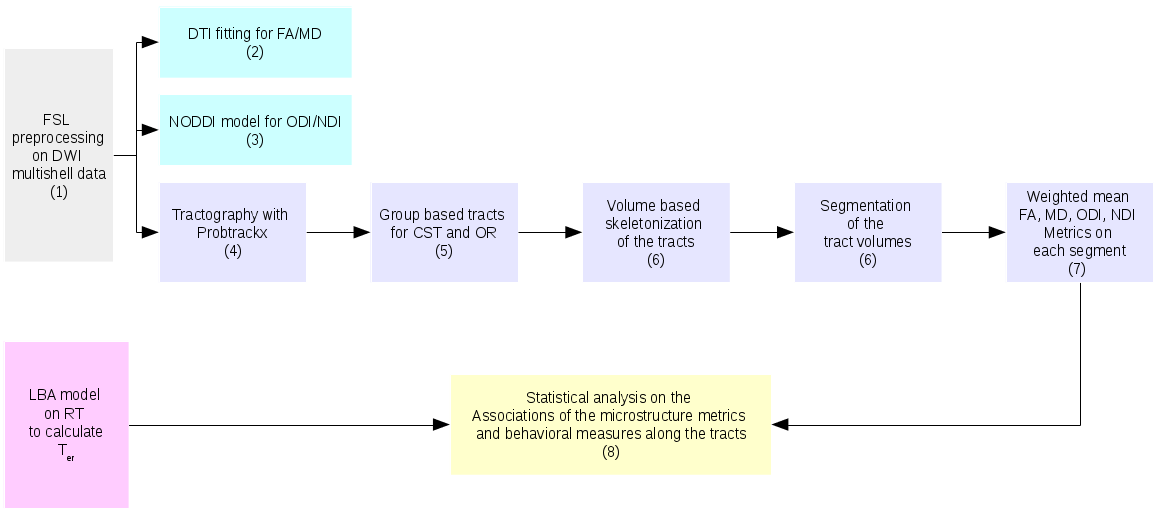
The pipeline is shown as:
Following steps correspond to the order in the pipeline. All MATLAB functions and bash scripts are provided.
-
Preprocessing of DWI files
FSL tools (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FDT/UserGuide)
-
DTI tensor fitting
FSL tools (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FDT/UserGuide)
-
NODDI fitting
NODDI toolbox (http://mig.cs.ucl.ac.uk/index.php?n=Tutorial.NODDImatlab)
-
Probabilistic Tractography
- Seed, waypoint and target masks to generate Corticospinal tracts, Optic radiations and Cingulum Bundles are given in the ROI folder. The scripts to generate subject-soecific ROIs are given in the Bash folder.
-
Group mask
- call_createGroupProbImage.m
- createGroupProbImage.m
-
Skeletonization ./Skeleton director
-
Main_tractSkeleton.m
- removeVoxel.m
- pruneBranchSkel.m
- organizeSkelLine.m
- spline3dCurveInterpolation.m
- limitSkelwithImage.m
- divideSkel.m
- assignVoxel2Skeleton.m
-
VSK toolbox (http://coewww.rutgers.edu/www2/vizlab/liliu/research/skeleton/VSK)
additional functions for the VSK
- findFirstandLastPointsTract.m
- findBoxVolume.m
- findImageCenter.m
modified functions of the VSK
- skel_Distmethod_mod.m
- skel_thinningmethod_mod.m
-
resampleSkeleton.m
- bordersWayPointROI_CST.m
- divideSkelOverlap.m
- assignVoxel2SkeletonOverlap.m
- writeLabelImageOverlap.m
- euclideanDist3.m
-
Visualization functions
- visualizeObject.m
- visualizeObjectSkel.m
- Individual Mapping of DTI and NODDI metrics ./TractStat Directory
-
Erode the FA maps
tbss_1_preproc (FSL)
-
Create WM masks from T1 images of subjects and register to native DWI space (b0) so that the FA and MD can be masked.
- segment_T1.sh
- register_WM_to_b0.sh
-
Register the tracts (fdt) back to the native DWI space
- register_fdt_to_b0.sh
-
Main_tractSkeletonStatFA.m
-
Main_tractSkeletonStatNODDI.m
- averageFAMDtractSkeletonOverlapReg.m
- saveAlongTractMetrixTxt.m
- plotMetricsAlongTract.m
-
Main_tractSkeletonStatWholeTract.m
- averagetract.m
- Statistics ./TractStat Directory
- statFunctList
-
statPalmImageCluster_CSV_TFCE.m
-
statPalmImageCluster_findSignificant.m
- plotRegTract.m
-
statMapCorrSkel2GroupIm.m
Note that: Any image viewer can be used to visualize the significant segments of the tracts - findIgnoranceColumns.m
-
-Partial Correlation on the whole tract
- statPartialCorrWholeTract.m
Dependency on other toolboxes:
- FSL (https://fsl.fmrib.ox.ac.uk/fsl)
- SPM12 (https://www.fil.ion.ucl.ac.uk/spm/software/spm12/)
- NODDI toolbox (http://mig.cs.ucl.ac.uk/index.php?n=Tutorial.NODDImatlab)
- VSK - *3 functions are modified (http://coewww.rutgers.edu/www2/vizlab/liliu/research/skeleton/VSK)
- splinefit (https://www.mathworks.com/matlabcentral/fileexchange/13812-splinefit)
- PALM (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/PALM)
- NaN (http://pub.ist.ac.at/~schloegl/matlab/NaN/)
- Gramm (https://www.mathworks.com/matlabcentral/fileexchange/54465-gramm-complete-data-visualization-toolbox-ggplot2-r-like)
- Shaded Error Bar (https://www.mathworks.com/matlabcentral/fileexchange/26311-raacampbell-shadederrorbar)
Auxiliary functions:
- takemeanNan.m
- takesemNan.m
- bootciMeanNan.m
Change the following files according to your environment:
- Set the root path in 'setDirectory.m'
- Run setEnvironment.m
- Run call_createGroupProbImage