This is a tentative experiments on solving automated neuron segmentation task using deep learing methods, residual networks.
See the original challenge post at: https://cremi.org, and leaderboard at: https://cremi.org/leaderboard/
Use Train.py file o train the models.
Best at 100 epoch:
Acc: 98.88%;
Loss: 0.0298
-
For this task, I trained a 2 way classifier to classify the central pixel in 127*127 sample as boudary and non-boundary. The 2-way sofmax layer was applied before the output of the network.
-
Reproduced and used residual network method. (original: https://arxiv.org/abs/1512.03385, implementation on github: https://github.com/gcr/torch-residual-networks). This has been giving me a great boost in classificaiton results.
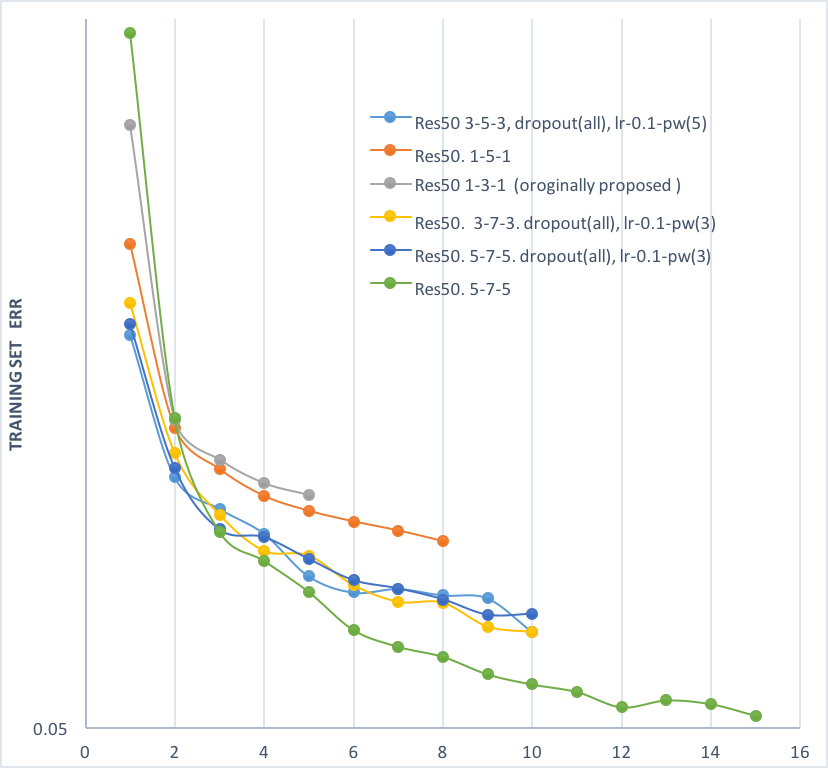
- (see plot below) It was found in preliminary experiments that using a 5-7-5 window for the three conv layers in the bottleneck block of residual net (training on 127*127 sample size, green line) outformed the originally proposed 1-3-1 structure (gray line) by a large margin, so experiments reported above were all trained with the 5-7-5. The position of batch normalization and dropout layer in the block was also changed.
- Selectively choose training samples from raw (see figure below): the yellow area X3 dilated boundary pixels were avoided to be chosen, only green and purple (true boudary, background) pixels will be selected into training batches,.
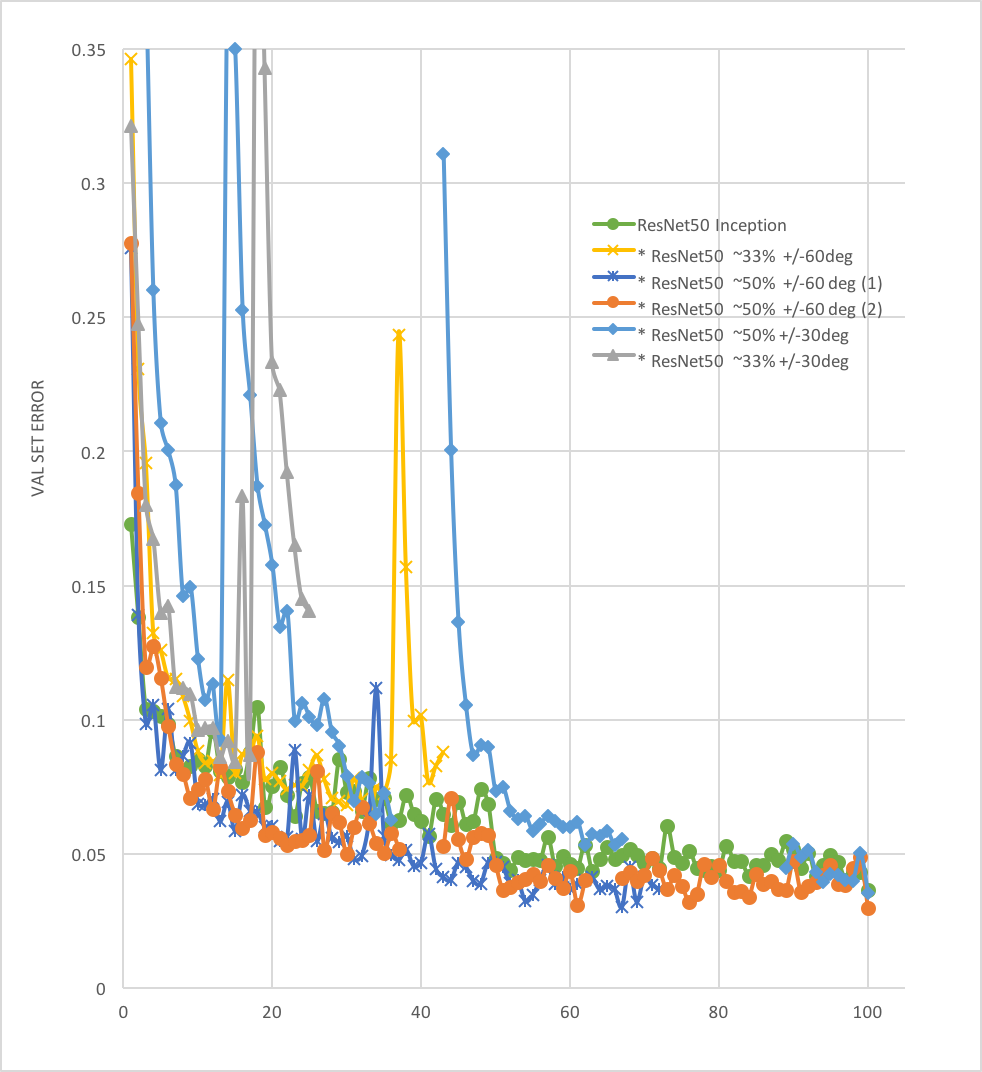
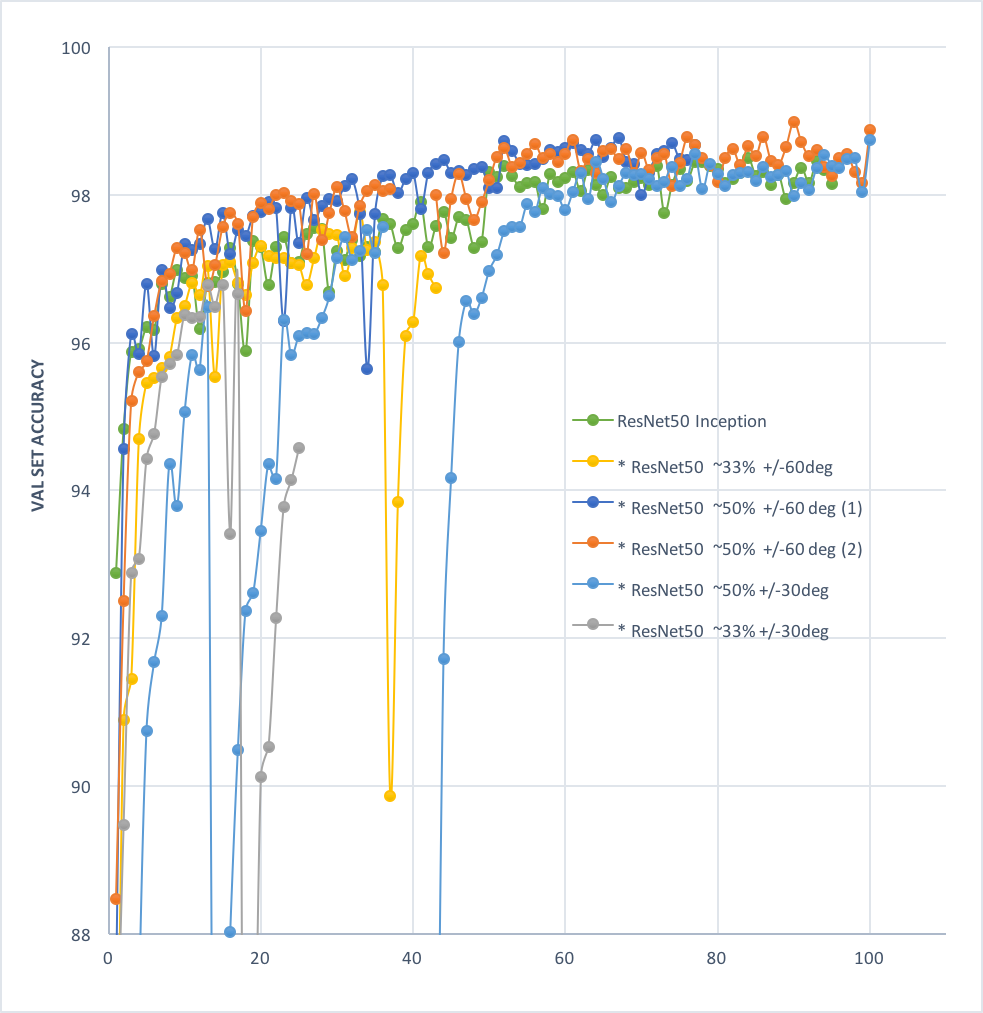
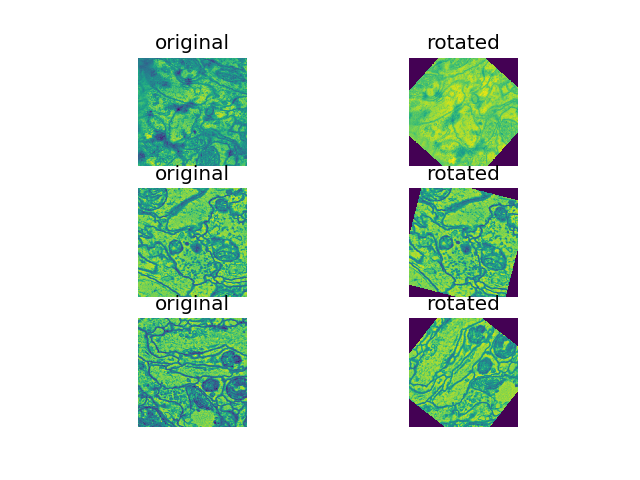
- Random rotation techniques: various augmentation approches were explored, including rand rotations of +/-60, rand +/- 30, on 33.33%, 50% of samples in each batc. rand +/- 60 deg on 50% of samples (see figure below) was found to perform the best so far.
- The neighbor area of the boundaries was avoided in this experiment, however the boundary pixels from other organels (intracellular organels) should also be avoided. These pixels could be easily treated as target neuron boundaries which are actually not. The approach to address this challenge can be to pre-train a network to recognize these intracellular boundaries and filter out these pixels when creating training batches for the segmentation task.
- the raw is originally a 3D image of size 125 * 1250 * 1250. I started by treating each layer in deapth 125 as an independent sample and trained my network with images in 2D sections. However, in later stages of experiments (which i was not able to do due to the time limit of my project), the third deimension should be considered to address the correlation between the neuron pixels at depth.
- python
- pytorch
- numpy
- matplotlib