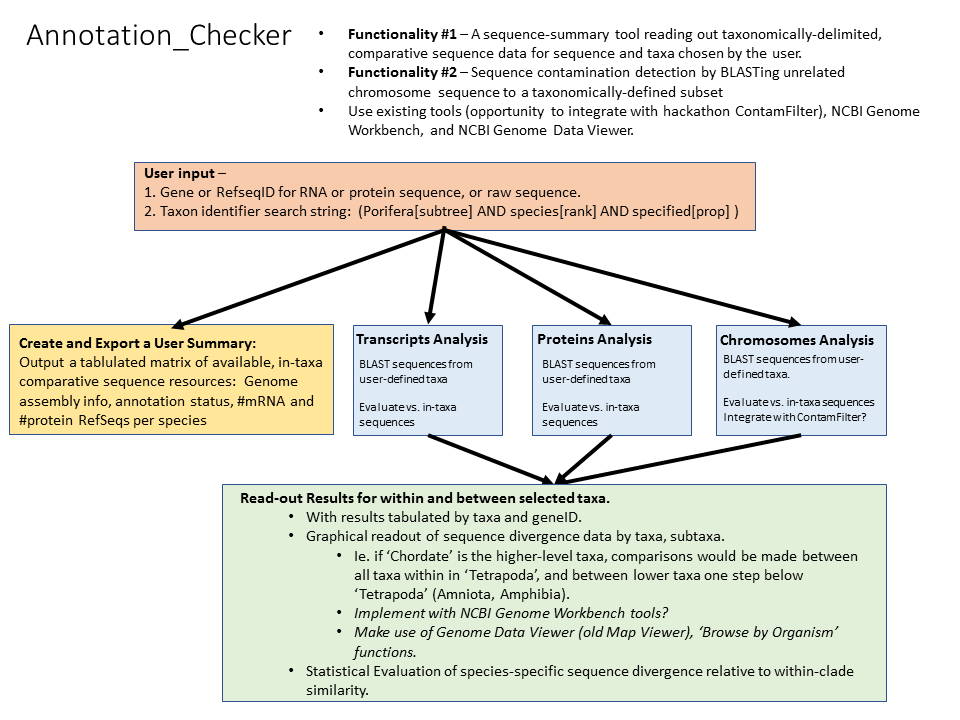
Annotation_Checker will provide a user-selectable, taxonomically subsetted, NCBI database-specific readout for identifying divergent taxonomic sequences. The tool will provide a quantitative capability to differentiate genome sequence quality from biologically-based evolutionary divergence.
20171125
$ esearch -db taxonomy -query "txid7215[Organism]" | elink -target assembly | efetch -format docsum | xtract -pattern DocumentSummary -element FtpPath_RefSeq | awk -F"/" '{print $0"/"$NF"_genomic.fna.gz"}'
20171121
$ wget `esearch -db genome -query txid10088[Organism:exp] | elink -target assembly | esummary | xtract -pattern DocumentSummary -element FtpPath_RefSeq | awk -F"/" '{print $0"/"$NF"_genomic.fna.gz"}'`
##downloads 3 .gz files, one for each refseq-listed genome:
GCF_000001635.26_GRCm38.p6_genomic.fna.gz GCF_900095145.1_PAHARI_EIJ_v1.1_genomic.fna.gz
GCF_900094665.1_CAROLI_EIJ_v1.1_genomic.fna.gz
$ esearch -db genome -query 53616[uid] | elink -target assembly | esummary | xtract -pattern DocumentSummary -element FtpPath_RefSeq
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/900/095/145/GCF_900095145.1_PAHARI_EIJ_v1.1
$ esearch -db genome -query txid40674[Organism:exp] | elink -target assembly | esummary | xtract -pattern DocumentSummary -element Organism SpeciesName FtpPath_RefSeq > Mammals
## Returns a list of Mammal species present in the Assembly DB, and ftp paths for Genomic fastas for those species with Genome RefSeq accessions.
NCBI Genomes Download (FTP) FAQ: https://www.ncbi.nlm.nih.gov/genome/doc/ftpfaq/
EUtils 'stuff': https://dataguide.nlm.nih.gov/eutilities/utilities.html
NCBI Assembly DB: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4702866/
Assembly DB Basics: https://www.ncbi.nlm.nih.gov/assembly/basics/
20171120
Get the four genome .gz files associated with Genome DB (esearch -db genome -query txid10088[Organism:exp]) from NCBI FTP site.
NCBI EUtils API tutorial: https://www.youtube.com/watch?v=iCFVVexp30o.
$ esearch -db genome -query txid10088[Organism:exp] | elink -target assembly | efetch -format uid
##Gives 25 genome assemblies ( = 4x species assembled mouse genomes (Mus musculus (22x), M. pahari(1x), M. caroli(1x), M. spretus(1x)),
##~Choose only 1 M.musculus assembly (Reference sequence).
1198761
1086041
1086031
738491
738481
738461
738451
731351
731341
731331
731321
731311
731301
731291
731281
731271
731261
731251
731241
663298
567298
419498
280718
239318
228898
$ esearch -db genome -query txid10088[Organism:exp] | efetch -format uid
##Gives 4 genome accessions (4x species assembled mouse genomes)
53616
34289
12818
52
$ esearch -db assembly -query txid10088[Organism:exp] | efetch -format uid
##Gives 40 assemblies
1198761
1086041
1086031
1034061
1033621
763931
738491
738481
738461
738451
731351
731341
731331
731321
731311
731301
731291
731281
731271
731261
731251
731241
663298
567298
558528
419498
418928
327618
315421
280718
239318
238988
228898
165668
132211
5728
5678
3188
3168
2598
ToDo
-mkblastdatabase from esearch results X, BLAST search against Taxa-selected assemblies from NCBI Assembly DB. Produce readouts of BLAST results.
-Alternatives for NCBI Taxonomy DB parsing: NCBI-Hackathons/EDirectCookbook#25
-VecScreen_plus_taxonomy, https://github.com/aaschaffer/vecscreen_plus_taxonomy
NCBI Entrez Tools (E-Utils) Manual: https://www.ncbi.nlm.nih.gov/books/NBK25501/pdf/Bookshelf_NBK25501.pdf
BLAST Command Line Applications Manual: https://www.ncbi.nlm.nih.gov/books/NBK279690/pdf/Bookshelf_NBK279690.pdf
20171117
$ esearch -db genome -query txid10088[Organism:exp] | esummary
$<!DOCTYPE DocumentSummarySet PUBLIC "-//NLM//DTD esummary genome 20140703//EN" "https://eutils.ncbi.nlm.nih.gov/eutils/dtd/20140703/esummary_genome.dtd">
<DocumentSummarySet status="OK">
<DbBuild>Build171117-1300.1</DbBuild>
<DocumentSummary><Id>53616</Id>
<Organism_Name>Mus pahari</Organism_Name>
<Organism_Kingdom>Eukaryota</Organism_Kingdom>
<Organism_Group></Organism_Group>
<Organism_Subgroup>Mammals</Organism_Subgroup>
<Defline></Defline>
<ProjectID>0</ProjectID>
<Project_Accession></Project_Accession>
<Status>Complete</Status>
<Number_of_Chromosomes>24</Number_of_Chromosomes>
<Number_of_Plasmids>0</Number_of_Plasmids>
<Number_of_Organelles>0</Number_of_Organelles>
<Assembly_Name>PAHARI_EIJ_v1.1</Assembly_Name>
<Assembly_Accession>GCA_900095145.2</Assembly_Accession>
<AssemblyID>1086041</AssemblyID>
<Create_Date>2017/01/10 00:00</Create_Date>
<Options></Options>
<Weight>0</Weight>
<Chromosome_assemblies>1</Chromosome_assemblies>
<Scaffold_assemblies>0</Scaffold_assemblies>
<SRA_genomes>0</SRA_genomes>
<TaxId>10093</TaxId>
</DocumentSummary>
<DocumentSummary><Id>34289</Id>
<Organism_Name>Mus caroli</Organism_Name>
<Organism_Kingdom>Eukaryota</Organism_Kingdom>
<Organism_Group></Organism_Group>
<Organism_Subgroup>Mammals</Organism_Subgroup>
<Defline>Mus caroli RefSeq Genome</Defline>
<ProjectID>264557</ProjectID>
<Project_Accession>PRJNA264557</Project_Accession>
<Status>Complete</Status>
<Number_of_Chromosomes>20</Number_of_Chromosomes>
<Number_of_Plasmids>0</Number_of_Plasmids>
<Number_of_Organelles>1</Number_of_Organelles>
<Assembly_Name>CAROLI_EIJ_v1.1</Assembly_Name>
<Assembly_Accession>GCA_900094665.2</Assembly_Accession>
<AssemblyID>1086031</AssemblyID>
<Create_Date>2017/01/10 00:00</Create_Date>
<Options></Options>
<Weight>0</Weight>
<Chromosome_assemblies>1</Chromosome_assemblies>
<Scaffold_assemblies>0</Scaffold_assemblies>
<SRA_genomes>0</SRA_genomes>
<TaxId>10089</TaxId>
</DocumentSummary>
<DocumentSummary><Id>12818</Id>
<Organism_Name>Mus spretus</Organism_Name>
<Organism_Kingdom>Eukaryota</Organism_Kingdom>
<Organism_Group></Organism_Group>
<Organism_Subgroup>Mammals</Organism_Subgroup>
<Defline>Mus spretus overview</Defline>
<ProjectID>83951</ProjectID>
<Project_Accession>PRJNA83951</Project_Accession>
<Status>Complete</Status>
<Number_of_Chromosomes>20</Number_of_Chromosomes>
<Number_of_Plasmids>0</Number_of_Plasmids>
<Number_of_Organelles>1</Number_of_Organelles>
<Assembly_Name>SPRET_EiJ_v1</Assembly_Name>
<Assembly_Accession>GCA_001624865.1</Assembly_Accession>
<AssemblyID>731341</AssemblyID>
<Create_Date>2016/04/15 00:00</Create_Date>
<Options></Options>
<Weight>0</Weight>
<Chromosome_assemblies>1</Chromosome_assemblies>
<Scaffold_assemblies>0</Scaffold_assemblies>
<SRA_genomes>0</SRA_genomes>
<TaxId>10096</TaxId>
</DocumentSummary>
<DocumentSummary><Id>52</Id>
<Organism_Name>Mus musculus</Organism_Name>
<Organism_Kingdom>Eukaryota</Organism_Kingdom>
<Organism_Group></Organism_Group>
<Organism_Subgroup>Mammals</Organism_Subgroup>
<Defline>The laboratory mouse is a major model organism for basic mammalian biology, human disease, and genome evolution, and its genome has been sequenced</Defline>
<ProjectID>9559</ProjectID>
<Project_Accession>PRJNA9559</Project_Accession>
<Status>Complete</Status>
<Number_of_Chromosomes>21</Number_of_Chromosomes>
<Number_of_Plasmids>0</Number_of_Plasmids>
<Number_of_Organelles>1</Number_of_Organelles>
<Assembly_Name>GRCm38.p6</Assembly_Name>
<Assembly_Accession>GCA_000001635.8</Assembly_Accession>
<AssemblyID>1198761</AssemblyID>
<Create_Date>2002/05/15 00:00</Create_Date>
<Options></Options>
<Weight>999</Weight>
<Chromosome_assemblies>17</Chromosome_assemblies>
<Scaffold_assemblies>5</Scaffold_assemblies>
<SRA_genomes>0</SRA_genomes>
<TaxId>10090</TaxId>
</DocumentSummary>
</DocumentSummarySet>
$ python3.6 Taxpull.py > taxids
##Taxpull.py
from ete3 import NCBITaxa
ncbi = NCBITaxa()
descendants = ncbi.get_descendant_taxa('Mus')
print(descendants)
$ [10089, 10091, 10092, 35531, 39442, 46456, 57486, 80274, 116058, 179238, 477815, 477816, 947985, 1266728, 1385377, 1643390, 1879032, 10096, 135827
, 135828, 10098, 270352, 270353, 10103, 27681, 83773, 186193, 186842, 254704, 473865, 481680, 10093, 41269, 10102, 10105, 41270, 60742, 273921, 27
3922, 397330, 397331, 397332, 397333, 544437, 887131, 1380868, 1582499, 1582501, 1582502, 1582503, 1582508, 1582509, 1890198, 10094, 10101, 210726
, 273920, 10095, 10108, 100383, 229288, 270354, 310402, 390847, 390848, 468371, 592722, 1390222, 1582479, 1582480, 1582481, 1582482, 1582483, 1582
504, 1582505, 1582506, 1582507]
20171115
-
Pull user-specified RefSeq genomes by NCBI Taxonomy Database taxID's (https://www.ncbi.nlm.nih.gov/taxonomy). ie. (Danio rerio).
-
Generate a list of taxID's from a given node (eg. taxID's for all formal species descending from 'Protostomia'.
-
Best tools available from Python-based ETE-Toolkit (http://etetoolkit.org/) , specifically the getting_descendant_taxa() command (http://etetoolkit.org/docs/2.3/tutorial/tutorial_ncbitaxonomy.html#getting-descendant-taxa), no quick alternatives appear elsewhere - may avoid "traversing the taxonomy database": (https://www.biostars.org/p/6528/) (https://www.biostars.org/p/16262/) (https://www.biostars.org/p/13452/)
Jaime Huerta-Cepas, Francois Serra and Peer Bork. ETE 3: Reconstruction, analysis and visualization of phylogenomic data. Mol Biol Evol 2016; doi: 10.1093/molbev/msw046
- Install ETE-toolkit. Windows-based Oracle virtual box or windows python?
11-17. Check each taxid for sequence database resources - 0/1? genomic DNA, RNA, protein databases, #accessions. Parse a subset of taxids possessing each resource.
20171114 -Began to work on user taxon and sequence selection.
-
Write a search string using NCBI EUtils to pull a reference sequence. NP_037367 = ribonuclease 3 isoform 1 [Homo sapiens]. https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=protein&term=NP_037367
-
Fetch the sequence fasta file. https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=protein&id=155030234&rettype=fasta&retmode=text
-
BLAST the reference fasta against user-selected taxID list. For testing and comparison, a list of 6 vertebrates (plus human reference) with well-validated genomic annotations, were chosen (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi). Each taxa was BLASTed against the user reference sequence, and the best hit was selected if BLAST result was valid:
taxIDs: 9913 - Bos taurus (domesticated cow) 7955 - Danio (zebrafish) 9606 - Human 10090 - Mouse 4932 - Rat 8355 - Takifugu (pufferfish) 4577 - Xenopus
- Continue working to automate this script.
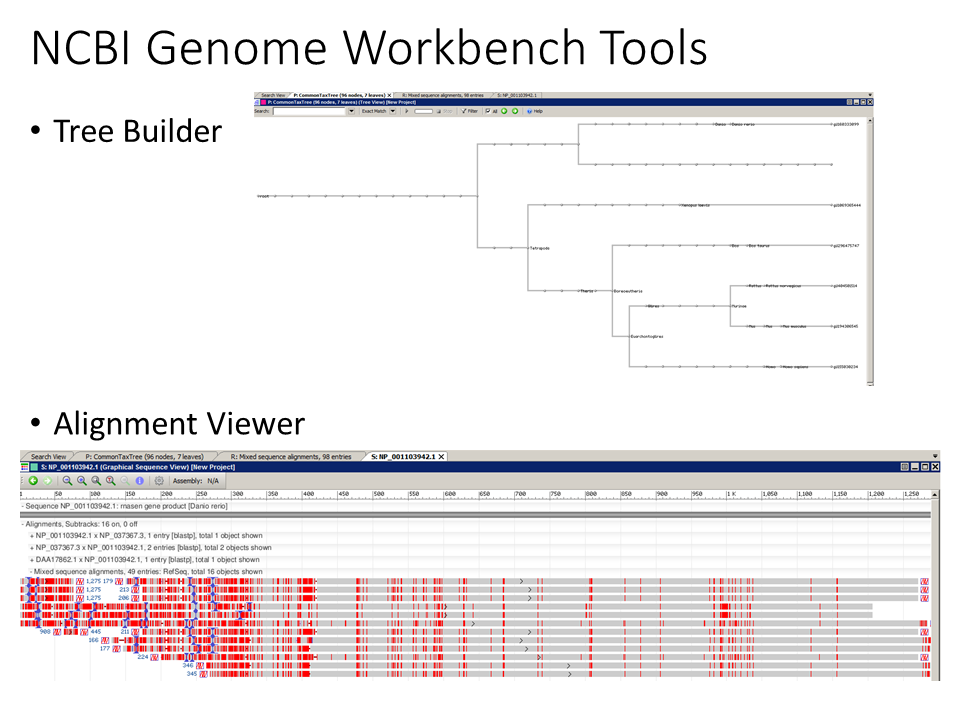
-NCBI Genome Workbench (https://www.ncbi.nlm.nih.gov/tools/gbench/) contains useful functionality. Work to incorporate useful features into Annotation_Checker User Summary readouts.