The objective of this assignment is to analyze the effect of gaps and number of species in efficiency of detecting relaxed selection by RELAX in terms of :
- number of gaps
- length of gaps
- number of species
Relaxed selection is a selective phenomena that occurs when selective pressures are either eliminated or reduced.
- Biotic sources :
- Predation elimination
- Elimination of pathogen
- Abiotic sources :
- Changes in light, temperature or water
- Changes composition of soil or mineral
Selection of any form (balancing, directional, etc.) can be relaxed. An example from Lahti et al. 2009 is here.
A hypothetical environmental change from an ancestral condition : possible outcomes -> (a) to (e)
We can detect signatures of selection from DNA sequence of organisms. The past fifty six years have seen the development and application of numerous statistical methods to identify genomic regions that appear to be shaped by natural selection. Natural selection is based on the simple observation of fitness-enhancing traits.
The way in which selection become observable and quantifiable.
Given that selection operates at the level of the phenotype, alleles showing evidence of selection are likely to be of functional relevance. There are several approaches available to detect selection at macroevolutionary scale.
What these methods does is :
- identify sequences that are likely to be functional (coding or conserved)
- Then search for lineage-specific accelerations in the rate of evolution.
Such accelerations are indicated by an excess of substitutions relative to the baseline mutation rate, which can be calculated from the number and rate of synonymous mutations.
Here we used a general hypothesis testing framework called RELAX from Hyphy package. HyPhy (Hypothesis Testing using Phylogenies) is an open-source software package for the analysis of genetic sequences for inferring natural selection using techniques of :
- phylogenetics
- molecular evolution
- machine learning
HyPhy distributes a variety of methods for inferring the strength of natural selection from the genetic data. In the case of branch-based methods for detecting selection, there is Relax.
The Decision tree : to find the appropriate method for detecting the molecular process of interest.
RELAX is a hypothesis testing framework that asks whether the strength of natural selection has been relaxed or intensified along a specified set of test branches.
Here the objective is to analyze the effect of gaps in detecting relaxed selection. Essentially, a gap occurs if something happens in our genome that can't be explained by uniformity and is also more than just mis-sequencing.
Here are some types of genome assembly gaps from Chaisson et al., 2015 :
(a) Sequence-coverage gaps: absence or reduction in sequence reads at that location.
(b) Segmental duplication-associated gaps: high sequence identity make read overlaps ambiguous.
(c) Satellite-associated gaps: higher-order tandem arrays of repetitive sequence cause read 'pileups'.
(d) Muted gaps: Contracted assembly relative to true genome.
One of the first problems anyone who do sequencing have to tackle is to distinguish the gap source between sequencing or alignment error versus actual indel in DNA. In such cases we have to minimize the false positives (type 1 error) and false negatives (type 2 error).
The presence of gaps can lead to several problems and ambiguities in assembly or alignment and hence the downstream analysis. These could lead to misinterpretation of the biology of data we are analyzing. As a matter of fact, here we try to analyze how the presence of gaps affect a particular downstream analysis - inference of strength of Natural Selection.
Two approaches to do this:
(1) Using a gene known to be under relaxed selection.
(2) Using simulated data.
In both cases we variably mask certain parts of the sequence as gaps and analyze the p values and k values inferred by Relax. To mask the sequence we used Bedtool's commands :
- random - generate a random set of intervals.
- maskfasta - masks sequences based on intervals.
How the bedtools 'random' and 'maskfasta' command works.
For the first approach we took a gene known to be under strong relaxed selection. Here we choose the gene CYP8B1 which is found and verified to be under strong relaxed selection in some mammals and birds (which come under a common clade called 'Amniota') by Shinde et al., 2019.
A small overview about CYP8B1 gene and protein
CYP8B1 is a single exonic gene that determines the ratio of primary bile salts. The loss of this gene has been linked to lack of cholic acid in naked mole rats, elephants and manatees. The Sagar et., 2019 used CYP8B1 gene ORFs from more than 200 species of birds and mammals to look for signatures of relaxed selection.
The taxonomic orders in Sagar et al., 2019 study are boxed red ~ 15 groups.
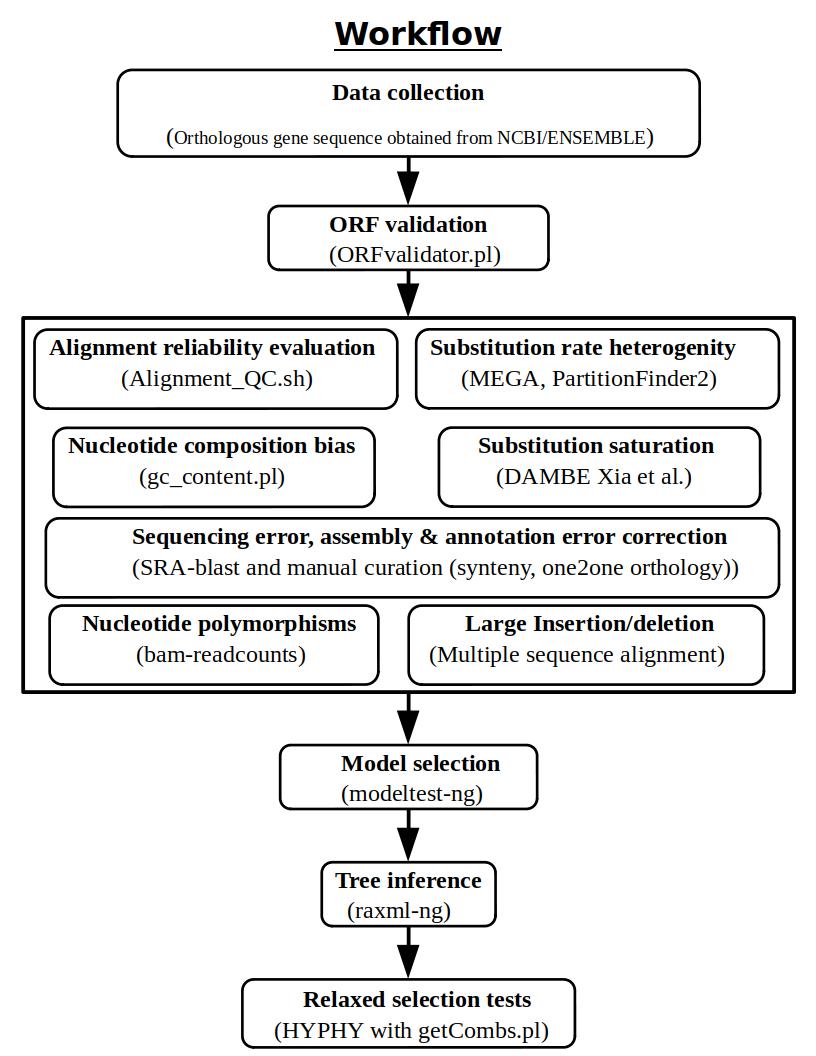
The test for the relaxed selection of CYP8B1 gene in the amniotes is carried out as per the pipeline mentioned in the Shinde et al., 2019. The major steps used in detection of the relaxed selection are listed below and a more detailed information is given in projects.
&nsbp;