Under development Deep learning library based on deep forests
A deep forest consists of a stack of layers, where each of them learns from both the input features and the output features of the previous layer. Each layer is made of a series of complete-random forests. This results in a whole hierarchy of predictors, from single trees to deep forests.
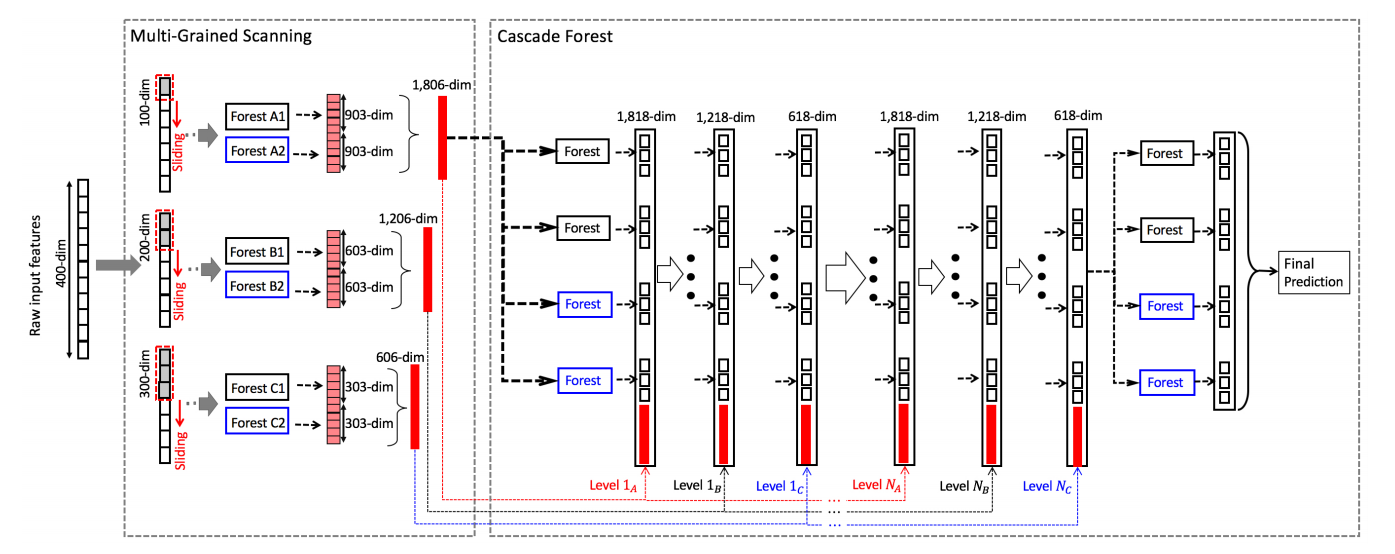
The rationale behind this project is to solve the problem of high dimensionality of fine grained scanners. Indeed, contrary to neural networks' convolutional layers, multi-grained scanners tend to increase the size of the temporary data due to slicing. To get around this issue, the concept of virtual datasets has been developed. Furthermore, fast tree learning algorithms have been implemented.
The idea is partly inspired by the original deep forest article:
Deep Forest: Towards An Alternative to Deep Neural Networks, by Zhi-Hua Zhou and Ji Feng (arXiv:1702.08835 [cs.LG])
From the root, build the library by taping the following commands:
$ cd python
$ sudo python setup.py installFitting a single tree
from scythe.core import *
config = TreeConfiguration()
config.min_threshold = 1e-06
config.max_height = 50
config.n_classes = 3
config.max_nodes = 30
config.nan_value = -1.0
tree = Tree(config, "classification")
tree.fit(X_train, y_train)
probas = tree.predict(X_test)Fitting a random forest:
fconfig = ForestConfiguration()
fconfig.n_classes = 2
fconfig.max_n_trees = 500
fconfig.bagging_fraction = 0.8
fconfig.max_depth = 50
fconfig.max_n_features = 20
forest = Forest(fconfig, "classification", "rf")
forest.fit(X_train, y_train)
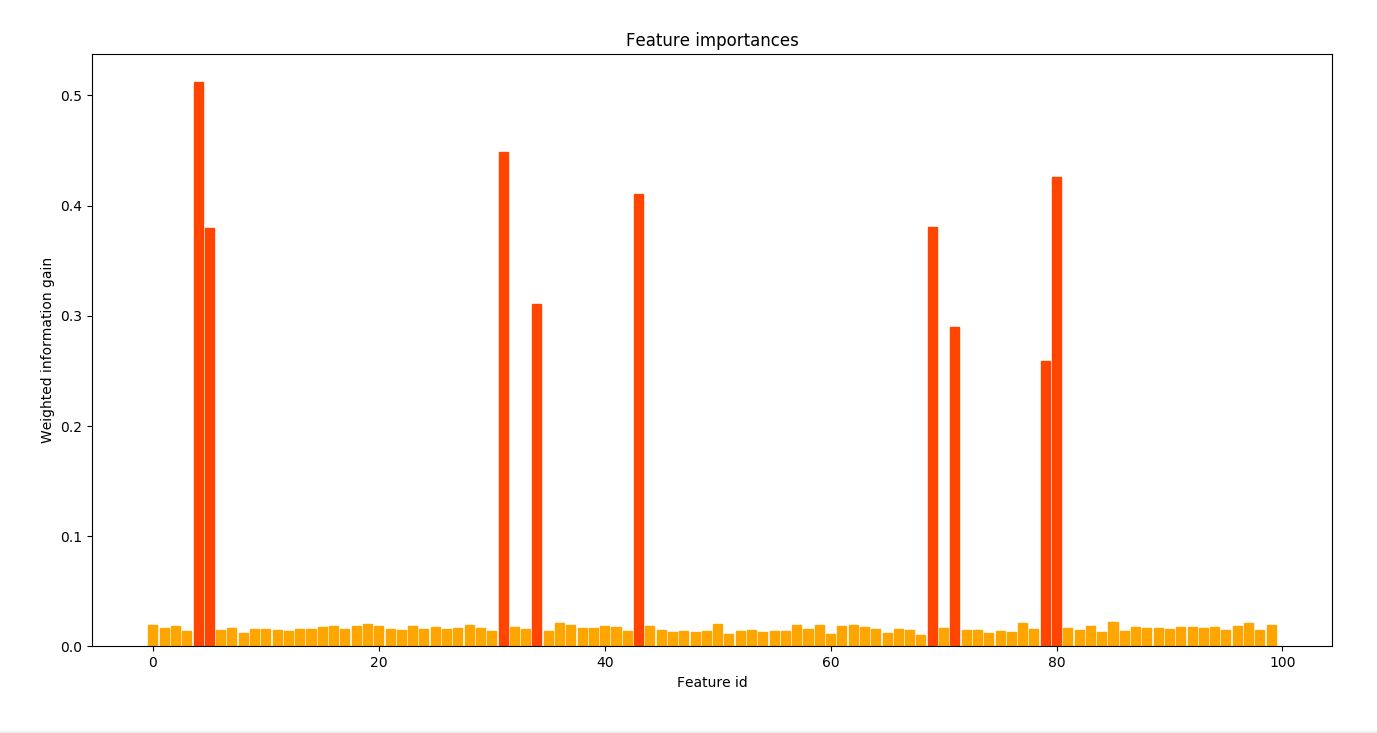
probas = forest.predict(X_test)Get the feature importances:
from scythe.plot import plot_feature_importances
plot_feature_importances(forest, alpha = 1.4)
plt.show()A complete example on the MNIST dataset can be found here.
Create an empty deep forest model:
from scythe.core import *
graph = DeepForest(task = "classification", n_classes = 10)Create a forest configuration:
from scythe.layers import *
fconfig = ForestConfiguration()
fconfig.bag_size = 60000
fconfig.n_classes = 10
fconfig.max_n_trees = 4
fconfig.max_n_features = 20
fconfig.max_depth = 12
lconfig = LayerConfiguration(fconfig, n_forests_per_layer, COMPLETE_RANDOM_FOREST)Add layers to model:
scanner = graph.add(MultiGrainedScanner2D(lconfig, (15, 15)))
cascade = graph.add(CascadeLayer(lconfig))
cascade2 = graph.add(CascadeLayer(lconfig))
cascade3 = graph.add(CascadeLayer(lconfig))Two layers are called connected if one is parent of the other and vice versa. Each new layer is automatically connected as a child of the rear layer. The rear layer is always the last cascade layer that has been added to the graph. To connect a layer to a non-rear layer, use the DeepForest.connect(parent_id, child_id) method.
graph.connect(scanner, cascade2)
graph.connect(scanner, cascade3)Then fit the model:
graph.fit(X_train, y_train)And finally predict on new instances:
probas = graph.classify(X_test)-
Create a R wrapper
-
Handle sparse matrices and arrays
-
Use float32 data samples instead of doubles
-
Skip the evaluation of data samples that already fell to the left of the previous split value (CART)
-
Avoid additional split values when evaluating a feature (save counters in temporary variables)
-
Design a heuristic for skipping split values that are not promising
-
Select a subset of features when selecting a split (random forests)
-
Set the minimum number of instances per leaf as a parameter (CART)
-
Use gradient boosting ?
-
Adapt random forests code for regression tasks
-
Proper setup.py file
-
Python wrapper of the LayerConfig structure
-
Sort queue by information gain in decision tree algorithm
Reference: http://web.mit.edu/insong/www/pdf/rpackage_instructions.pdf